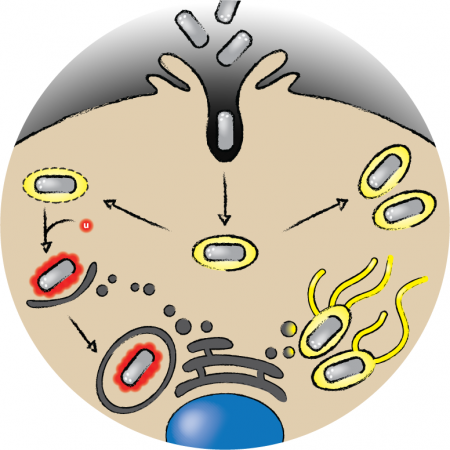
Bacterial resistance to standard antibiotic treatment has evolved into a serious threat worldwide. We believe that deciphering pathogenic and host defense mechanisms during bacterial infections is the key to novel antibacterial strategies. Ubiquitin (Ub) drives innate immune receptor signaling as well as microbicidal programs including selective autophagy of intracellular pathogens (Gomes and Dikic, 2014). Although prokaryotes lack Ub-coding genes as well as the Ub-proteasome system, a wide range of bacterial pathogens acquired strategies to exploit the host Ub machinery for their life cycle, counteract host inflammatory signaling and immune defense programs. In our group, we intensively study Legionella- and Salmonella-induced cell response. We and others have demonstrated that intracellular ubiquitinated bacteria are recognized by various glycan-, Ub- and LC3-binding autophagy receptors including p62/SQSTM1, NDP52 and LGALS8/Galectin 8, OPTN and TAX1BP1, which all help to deliver the pathogen into autophagosomes for degradation (Wild et al., 2011, van Wijk et al., 2012, van Wijk et al., 2017). Using our established quantitative diGly proteomics platform, we recorded the first global ubiquitinome of host epithelial cells infected with Salmonella typhimurium (Fiskin et al, 2016). We further acquired knowledge of the Salmonella infection approaching other available in the lab techniques. The Legionella study focusses on dissecting the role of Ser-Ub.
 STRUCTURE
STRUCTURE PROTEOMICS
PROTEOMICS MICROSCOPY
MICROSCOPY CANCER
CANCER ER-PHAGY
ER-PHAGY Ser-UB
Ser-UB