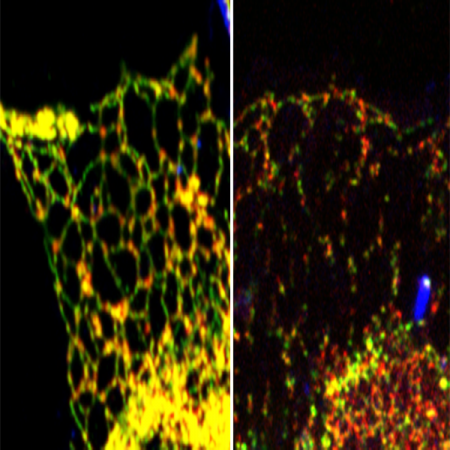
Legionella pneumophila is a gram negative bacteria that causes Legionnaires disease. Upon infection, the bacteria eject more than 300 effector proteins into the host, which then hijacks host cell metabolism for bacterial survival and proliferation in intracellular vacuoles (Legionella-containing vacuoles). One of the pathways that get reprogrammed by the bacteria is the cellular ubiquitination system. Legionella has a unique system of targeting the cellular ubiquitin pool by modifying ubiquitin by phosphoribosylation. This ubiquitin (PR-ubiquitin) is then transferred to host proteins by phosphoribose-linked serine ubiquitination.
Our lab focusses on dissecting the role of phosphoribose-linked serine ubiquitination using a combination of biochemistry, structural biology and mass spectrometry to discover the molecular basis of phosphoribose-linked ubiquitination catalyzed by SidE effector proteins. These are coupled to Legionella infection studies to understand the effect of bacterial effectors on cellular signaling pathways and organellar dynamics. PR-ubiquitination is a fine-tuned response and is regulated during infection using at least two other bacterial effectors – SidJ and Dups. From Cryo-EM studies and mass spectrometric signatures, we identified SidJ to be a bacterial glutamylase, which glutamylates and inactivates the active site of SidE proteins (Bhogaraju et al., 2019). Dups were also a recent serendipitous discovery and were characterized to be a group of deubiquitinases that can specifically reverse PR-ubiquitination (Shin et al., 2020). Using a catalytically dead Dup-trapping mutant, coupled to mass spectrometry, we have identified more than 180 cellular proteins that are modified by PR-ubiquitination during Legionella infection, showing a global effect of SidE effectors in reprogramming the host cell during infection.
 STRUCTURE
STRUCTURE PROTEOMICS
PROTEOMICS MICROSCOPY
MICROSCOPY BACTERIA
BACTERIA