Computational Cell Biology
Welcome to the Bhaskara lab.
Our team focuses on tackling challenging problems at the interface of Cell Biology, Physics, and, Data Sciences.
We develop and apply theoretical modeling, novel computer simulations, and, data integration methods to understand complex biological systems.
Integrative Modeling
We employ state-of-the-art bioinformatics and AI methods to decode relationships between protein sequence, structure, function & Evolution. We model the 3D organization of proteins, their functional interactions (biological complexes), and post-translational modifications by integrating structural data from various experimental and computational methods.
Biomolecular Simulations
We study biological pathways involving protein-induced membrane mechanics to reveal molecular mechanisms at cellular and organellar membranes (e.g. Autophagy pathways). Within these projects, we aim at capturing the dynamics of biomolecular systems at various temporal and spatial scales in quantitative terms to (i) provide mechanistic understanding & erect testable hypotheses, (ii) guide the design of new experiments, and (iii) provide a theoretical framework for experimental observations and simulations.
Data Integration
We employ data science approaches to integrate disparate datasets effectively. We combine information from large-scale HTP datasets (Proteomic, Genetic, Chemical, and imaging) with bioinformatic predictions and publicly available repositories to mine and learn underlying biological patterns.
Fellowships, Ph.D., and Postdoc positions
Please visit the IBCii Career page for details on current vacancies and open positions.
New Postdoc position to work on Molecular Mechanisms in Selective Autophagy.
We are always looking for motivated and skilled students to join our team and pursue internships, Bachelor's, and Master's thesis projects. Please contact us directly.
Projects for Master's theses, HiWi, and internships: Please contact Dr. Ram Bhaskara for these projects.
Dr. Ramachandra M Bhaskara
Ram Bhaskara received his bachelor's degree in Chemistry and Biochemistry from Osmania University, India. He then joined the Integrated Ph.D. program in Biological Sciences at the Indian Institute of Science, India. During this period, he gained research experience in various labs (from field ecology to molecular biophysics). He then joined Prof. N. Srinivasan's group for his Master's and Ph.D. During this period, he developed novel computational tools and analysis schemes to understand the "Structure, Stability, and Evolution of Multi-domain proteins." Ram was honored with the B. H. Iyer Gold Medal for the best doctoral thesis from the Molecular Biophysics Unit (2014). He then began his post-doctoral stint with Prof. Gerhard Hummer at the Max-Planck Institute of Biophysics, Frankfurt. Ram worked on diverse membrane remodeling processes: curvature induction, budding, fusion, & poration, primarily using physics-based modeling and simulation approaches. He developed and pioneered novel simulation methods to model dynamic processes inaccessible to experiments. Ram moved to the Institute of Biochemistry II, Goethe University, in September 2020 as a team leader for Computational Cell Biology to work on Data integration problems, modeling, and simulations of complex systems. He is interested in understanding biophysical mechanisms shaping the complex cellular architecture at various scales. His research focuses on developing computational tools to analyze and decipher molecular mechanisms.
Alberto Cristiani
Alberto Cristiani studied at the University of Trieste, Italy, and graduated with a degree in Nano-Biotechnology. For his bachelor’s thesis, he worked on identifying inhibitors of Pin1 at the LNCIB laboratory, Italy. He then switched fields to study Computational Biology by joining the Master’s program in Systems Biology at Newcastle University. During this period, he acquired an interdisciplinary skillset and an interest in the quantitative modeling of biological systems. As part of his thesis, he developed a method to automatically detect a set of functional Boolean Networks within the larger gene regulatory networks. After his Master’s degree, he worked briefly as a programmer (AI and Gameplay) at Symmetrical and later as an ETL developer in the Business Intelligence unit of NTT Data, Italy. Alberto then returned to academics and is now a Ph.D. student at the Computational Cell Biology group. He is currently developing pipelines for data acquisition and machine learning in order to explore the biology of the autophagy, ubiquitin, and SUMO systems.
Duy Nguyen
After finishing high school in Vietnam, Duy studied and graduated in Bachelor Biophysics at the University of Frankfurt am Main. Since then he is pursuing his Master's degree in Biochemistry, also from Goethe University Frankfurt am Main. His main focus is on Bioinformatics, Data Science and Autophagy. For his Master thesis, he is using data science approaches to identify novel autophagy receptors.
Izabela Dwojak
Izabela is a Master's student (Biophysics) at the Johann Wolfgang von Goethe University in Frankfurt. For her bachelor's thesis at the Institute of Atomic Physics, she studied simulations of strong-field ionization processes in three-dimensional laser fields. She is pursuing an internship in the CCB group and is interested in image-processing approaches to quantify organelle morphology. Currently, she is on an Erasmus exchange program in Italy.
Kateryna Lohachova
Kateryna Lohachova obtained both her Bachelor and Master degrees in Chemistry from V.N. Karazin Kharkiv National University under the supervision of Prof. Oleg Kalugin. She studied the electrochemical applications of binary non-aqueous systems based on 1:1 electrolytes, ionic liquids, and molecular liquids. Her research was dedicated to exploring the features of molecular dynamics modeling as a method for predicting the behavior of ion-molecular systems, as well as the peculiarities of transport properties and ionic aggregation in such systems. During the SARS-CoV-2 pandemic, she worked with Prof. Alexander Kyrychenko as part of Prof. Oleg Kalugin’s team, on a project dedicated to the molecular design and computer modeling of new inhibitors of the SARS-CoV-2 coronavirus. In April 2022, Kateryna joined the CCB team as a fellow of IBC2 first and since August 2023 as a PhD student under the supervision of Dr. Ramachandra M. Bhaskara, focusing on the membrane remodeling. Her main work is related to quantification of membrane curvature induced by membrane remodelling proteins.
Plamen Kondev
Plamen Kondev is currently pursuing his Master's degree in Biochemistry, Goethe University, Frankfurt. He was intern in the CCB group in 2021–2022. He pursued another internship in the Münch group and returned to the CCB group in Decmber 2024 to work on his MS theisis. He is interested in Reticulon proteins and how their membrane remodeling signatures.
Dr. Sergio Alejandro Poveda Cuevas
Alejandro received his bachelor's degree in biology from the Pedagogical and Technological University of Colombia. He continued his academic journey by earning a Master's in Bioinformatics from the University of São Paulo (USP), where he conducted research on cancer-related protein complexes and the refinement of force-field models for coarse-grained systems. He later pursued doctoral studies at USP, focusing his thesis on investigating the NS1 protein of the Zika virus. His research, utilizing molecular simulations, explored critical aspects such as protein oligomerization, membrane binding affinity, epitope estimation, and glycosylation, providing invaluable insights into molecular mechanisms and virulence of Zika virus. Currently, Alejandro is a post-doctoral researcher at the Computational Cell Biology lab, where he studies the biophysical properties of proteins involved in autophagy, with a particular emphasis on selective autophagy receptors. His primary research focuses on understanding the intrinsically disordered region (IDR) of the ER-phagy receptor FAM134B and its role in large-scale membrane remodeling events. His work revealed how the context-dependent, or "Janus-like" ensemble properties of the IDR actively influence membrane properties to both induce and sense positive membrane curvature, as well as facilitate the clustering and sorting of proteins during spontaneous budding processes.
Dr. Siddhanta Nikte
Siddhanta completed her Bachelor's degree (Biotechnology) from Fergusson College, Pune, India. She then pursued a Master's degree in Bioinformatics at the Bioinformatics Centre, Savitribai Phule Pune University (2016). For her Master's thesis, she worked with Dr. Manali Joshi to study the interactions of novel antimicrobial peptides with membranes. She then began her Ph.D. at the National Chemical Laboratory, Pune, in affiliation with the Academy of Scientific and Innovative Research, India under the guidance of Dr. Durba Sengupta. Her doctoral research focused on G-protein-coupled receptors (GPCRs), investigating how amino acid residues, dynamic domains, and lipid environments modulate receptor function. Following her Ph.D., she worked as a DBT-Research associate with Dr. Arnab Gupta at IISER-Kolkata, India, exploring complex aspects of membrane proteins for a brief period. Currently, Siddhanta is a postdoctoral researcher in the Computational Cell Biology (CCB) Lab, where her interests lie in the structural modeling, molecular docking, and simulation of E3 ligases, with a focus on their roles in cellular signaling and targeted protein degradation.
Past Members:
Dr. Arghya Dutta
Postdoc (2021–2024).
Arghya is currently an Assistant professor at the Department of Physics, SRM University, India.
Carolin Lange
BS Intern & BS Thesis (2021–2023).
Carolin is currently an Education Content Manager at Seneca Learning.
Dishant Loliyana
MS Intern (2025).
Dishant is currently finishing his Masters in Chemistry at the University of Siegen, Germany.
Jonathan Eisert
MS Intern (2021).
Jonathan moved to Merck Healthcare as an intern and is currently pursuing his PhD at TU Darmstadt.
Karling Tan
MS Intern (2024).
Karling Tan is currently finishing her Masters in Physical Biology of Cells and Cell Interactions at Goethe University Frankfurt.
Leo Jankovic
BS Intern (2024).
Leo Jankovic is currently completeing his undergraduate studies at Stanford University.
Our Publications
2025
- Buonomo, V., Lohachova, K., Reggio, A., Cano-Franco, S., Michele, C., Santorelli, L., Venditti, R., Polishchuk, E., Peluso, I., Brunello, L., Cirillo, C., Petrosino, S., Malan, S., De Cegli, R., Di Bartolomeo, S., Gargioli, C., Swuec, P., Cortese, M., Stolz, A., Bhaskara, R. M., Grumati, P. (2025) Two FAM134B isoforms differentially regulate ER dynamics during myogenesis. EMBO J. 1-35.
- Poveda-Cuevas, S. A., Lohachova, K., Markusic, B., Dikic, I., Hummer, G., Bhaskara, R. M. (2025) Janus-like behavior of intrinsically disordered regions in reticulophagy. Autophagy. 1-3 (Punctum).
2024
- Herhaus, L., Gestal-Mato, U., Eapen, V. V., Macincovik, I., Baily, H. J., Prieto-Garcia, C., Misra, M., Jacomin, A. C., Ammanath, A. V., Bagaric, I., Michaelis, J., Vollrath, J., Bhaskara, R. M., Bündgen, G., Covarrubias-Pinto A., Hunsjak, K., Zöllner, J., Gikandi, A., Ribicic, S., Bopp, T., van der Haeden van Noort, G. J., Langer, J. D, Weigert, A., Harper, W., Mancias, J. D., Dikic, I. (2024) IRGQ-mediated autophagy in MHC class I quality control promotes tumor immune evasion. Cell 187, 1-18.
- Poveda-Cuevas, S. A., Lohachova, K., Markusic, B., Dikic, I., Hummer, G., Bhaskara, R. M. (2024) Intrinsically disordered region amplifies membrane remodeling to augment selective ER-phagy. Proc. Natl. Acad. Sci. USA. 121 (44) e2408071121.
2023
- Mende, H., Khatri, A., Carolin, L., Poveda-Cuevas, S. A., Tascher, G., Covarrubias-Pinto, A., Löhr, F., Koschade, S.E., Dikic, I., Münch, C., Bremm, A., Brunetti, L., Brandts, C.H., Uckelmann, H., Dötsch, V., Rogov, V.V., Bhaskara, R. M. Müller, S. (2023) An atypical GABARAP binding module drives the pro-autophagic potential of the AML-associated NPM1c variant. Cell Reps. Apr 42, 113484.
- Foronda, H., Fu, Y., Covarrubias-Pinto, A., Bocket, H. T., González, A., Seemann, E., Franzka, P., Bock, A., Bhaskara, R. M., Liebmann, L., Hoffmann, M. E., Katona, I, Koch, N., Weis, J., Kurth, I., Gleeson, J. G., Reggiori, F., Hummer, G., Kessels, M. M., Qualmann, B., Mari M., Dikić, I., Hübner, C. A. (2023). Heteromeric clusters of ubiquitinated ER-shaping proteins drive ER-phagy. Nature 618, 394-401.
- González, A., Covarrubias-Pinto, A., Bhaskara, R. M., Glogger, M., Kuncha, S. K., Xavier, A., Seemann, E., Misra, M., Hoffmann, M. E., Bräuning, B., Balakrishnan, A., Qualmann, B., Dötsch, V., Schulman, B. A., Kessels, M. M., Hübner, C. A., Hummer, G., Dikić, I. (2023). Ubiquitin regulates ER-phagy and remodelling of endoplasmic reticulum. Nature 618, 394-401.
- Cristiani, A., Dutta, A., Poveda-Cuevas, S. A., Kern, A., Bhaskara, R. M. (2023) Identification of potential selective autophagy receptors from protein-content profiling of autophagosomes. J. Cell. Biochem. Apr 23. p1-14, Epub ahead of print PMID: 37087736.
2021
- Reggio A., Bunomo, V., Berkane, R., Bhaskara, R. M., Tellechea, M., Peluso, I., Polischuk, E., Di Lorenzo, G., Crillo, C., Esposito, M., Hussain, A., Huebner, A.K., Settembre, C., Hummer, G., Grumati, P., Stolz, A. (2021) Role of FAM134 paralogues in endoplasmic reticulum remodelling, ER-phagy and Collagen quality control EMBO Rep. 22: e52289.
- Ho, N. T., Siggel, M., Camacho, K. V., Bhaskara, R. M., Hicks, J. M., Yao, Y., Zhang, Y., Köfinger, J., Hummer, G., Noy, A. (2021) Membrane fusion and drug delivery with carbon nanotube porins. Proc. Natl. Acad. Sci. USA. 118, e2016974118.
- Siggel, M., Bhaskara, R. M., Moesser, M.K., Dikic, I., Hummer, G. (2021) FAM134B-RHD protein clustering drives spontaneous budding of asymmetric membranes. J. Phys. Chem. Lett. 12 (7), 1926–1931.
2020
- Vögele, M., Bhaskara, R. M., Mulvihill, E., van Pee, K., Yildiz, Ö., Kühlbrandt, W., Müller, D. J., Hummer, G. (2020) Reply to Desikan et al.: Micelle formation among various mechanisms of toxin pore formation. Proc. Natl. Acad. Sci. USA. 117 (10) 5109-5110.
- Herhaus, L., Bhaskara, R. M.†, Lystad, A. H.†, Simonsen, A, Hummer, G., Dikic, I. (2020) TBK1-mediated phosphorylation of LC3C and GABARAPL2 controls autophagosome shedding by ATG4 protease. EMBO Rep. 21: e48317
2019
- Siggel, M., Bhaskara, R.M., Hummer, G. (2019) Phospholipid scramblases remodel the shape of asymmetric membranes. J. Phys. Chem. Lett. 10 (20), 6351-6354.
- Vögele, M., Bhaskara, R.M., Mulvihill, E., van Pee, K., Yildiz, Ö., Kühlbrandt, W., Müller, D. J., Hummer, G. (2019) Membrane perforation by the pore-forming toxin pneumolysin. Proc. Natl. Acad. Sci. USA. 116 (27) 13352-13357.
- Bhaskara, R. M., Grumati, P., Garcia-Padro, J., Kalayil, S., Covarrubais-Pinto., A., Chen, W., Kudrayshev, M., Dikic, I., Hummer, G. (2019) Curvature induction and membrane remodelling by FAM134B reticulon homology domain. Nat. Commun. 10:2370.
2017
- D’Imprima, E., Salzer, R.†, Bhaskara, R. M.†, Sánchez R., Rose, I., Hummer, G., Kühlbrandt, W., Vonck, J., Averhoff, B. (2017) Cryo-EM structure of the bifunctional secretin complex of Thermus thermophilus. eLife 6: e30483.
- Bhaskara, R.M.†, Linker, S.M.†, Vöegele, M., Köfinger, J., Hummer, G. (2017) Carbon nanotubes mediate fusion of lipid vesicles. ACS Nano 11 (2), 1273-1280.
2016
- Ramachandran, R., Joseph, A. P., Bhaskara, R. M., Srinivasan, N. (2016) Structural and mechanistic insights into human splicing factor SF3b complex derived using an integrated approach guided by the cryo-EM density maps. RNA Biology 13 (10), 1025-1040.
2015
- Mehrotra, P., Bhaskara, R. M., Gnanavel, M., Rakshambikai, R., Martin, J., Srinivasan, N. (2015) Classification of protein sequences with special references to multi-domain systems. J. Biomol. Struct. Dyn. 33 (sup1), 110
- Craveur, P., Joseph, A. P., Esque, J., Narwani, T. J., Noël, F., Shinada, N., Goguet, M., Leonard, S., Poulain, P., Bertrand, O., Faure, G., Rebehmed, J., Ghozlane, A., Swapna, L. S., Bhaskara, R. M., Barnoud, J., Téletchéa, S., Jallu, V., Cerny, J., Schneider, B., Etchebest, C., Srinivasan, N., Gelly, J. G., de Brevern A. G. (2015) Protein flexibility in the light of structural alphabets. Front Mol. Biosci. 2 (20).
2014
- Bhaskara, R. M., Padhi, A., Srinivasan, N. (2014) Accurate prediction of interfacial residues in two-domain proteins using evolutionary information: implications for three-dimensional modeling. Proteins: Struct., Funct., Bioinf. 82 (7), 1219-1234.
- Gnanavel, M., Mehrotra, P., Rakshambikai, R., Martin, J., Srinivasan, N. Bhaskara, R. M. (2014) CLAP: A web-server for automatic classification of proteins with special reference to multi-domain proteins. BMC Bioinformatics.15 (343).
- Bhaskara, R. M., Mehrotra, P., Rakshambikai, R., Gnanavel, M., Martin, J., Srinivasan, N. (2014) The relationship between classification of multidomain proteins using an alignment-free approach and their functions: a case study with immunoglobulins. Mol Biosyst. 10 (5), 1082-1093.
2012
- Bhaskara, R. M., de Brevern, A. G., Srinivasan, N. (2012) Understanding the role of domain-domain linkers in the spatial orientation of domains in multi-domain proteins. J. Biomol. Struct. Dyn. 31(12), 1467-1480.
- Swapna, L.S., Bhaskara, R. M., Sharma, J., Srinivasan, N. (2012) Roles of residues in the interface of transient protein-protein complexes before complexation. Sci. Rep. 2, 334.
2011
- Bhaskara, R. M., and Srinivasan, N. (2011) Stability of domain structures in multi-domain proteins. Sci. Rep. 1, 40.
- Srinivasan, N., Agarwal, G., Bhaskara, R. M., Gadkari, R., Krishnadev, O., Lakshmi, B., Mahajan, S., Mohanty, S., Mudgal, R., Rakshambikai, R., Sandhya, S., Sudha, G., Swapna, L. S., Tyagi, N. (2011) Influence of genomic and other biological data sets in the understanding of protein structures, functions, and interactions. International Journal of Knowledge Discovery in Bioinformatics (IJKDB) 2, 24-44.
2010
- Indu, S†., Kumar, S.T†., Thakurela, S., Gupta, M., Bhaskara, R. M., Ramakrishnan, C., Varadarajan, R. (2010). Disulfide conformation and design at helix N-termini. Proteins: Struct., Funct., Bioinf. 78 (5), 1228-1242.
2009
- Bhaskara, R. M., Brijesh, C. M., Ahmed, S., Borges, R. M. (2009) Perception of ultraviolet light by crab spiders and its role in the selection of hunting sites. J. Comp. Physiol. A (Neuroethol. Sens. Neural. Behav. Physiol.) 195 (4), 409-417.
We employ diverse state-of-the-art bioinformatics, physics-based and AI methods to decode relationships between protein sequence, structure, function & Evolution. We model the 3D organization of proteins, their functional interactions (biological complexes), and post-translational modifications by integrating structural data from various experimental and computational methods.
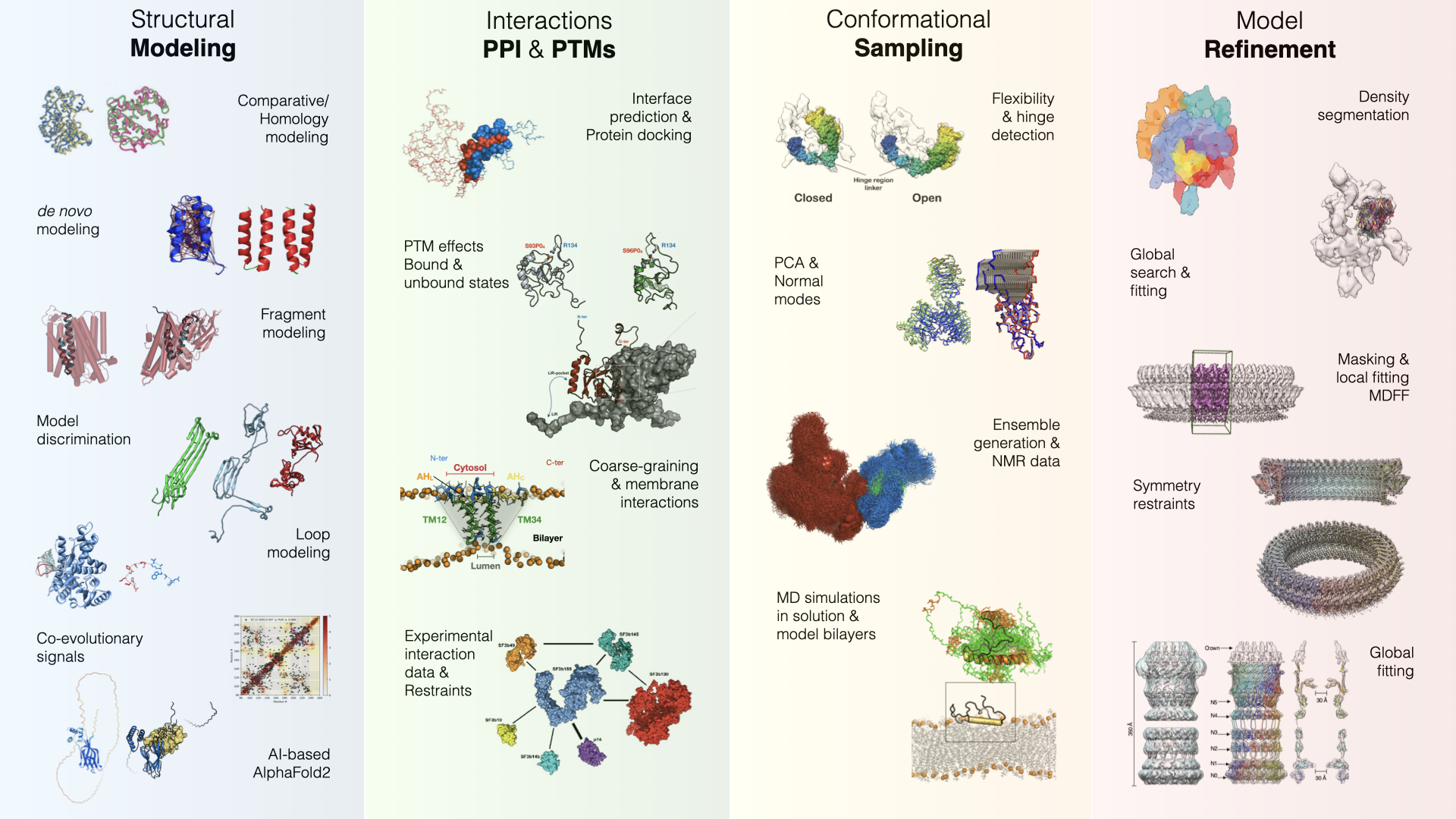
Selected publications
-
Herhaus, L., Bhaskara, R.M.†, Lystad, A.H.†, Simonsen, A, Hummer, G., Dikic, I. (2020) TBK1-mediated phosphorylation of LC3C and GABARAPL2 controls autophagosome shedding by ATG4 protease. EMBO Rep. 21: e48317
-
Vögele, M., Bhaskara, R.M., Mulvihill, E., van Pee, K., Yildiz, Ö., Kühlbrandt, W., Müller, D.J., Hummer, G. (2019) Membrane perforation by the pore-forming toxin pneumolysin. Proc. Natl. Acad. Sci. USA. 116 (27) 13352-13357.
-
Bhaskara, R.M., Grumati, P., Garcia-Padro, J., Kalayil, S., Covarrubais-Pinto., A., Chen, W., Kudrayshev, M., Dikic, I., Hummer, G. (2019) Curvature induction and membrane remodelling by FAM134B reticulon homology domain. Nat. Commun. 10:2370.
-
D’Imprima, E., Salzer, R.†, Bhaskara, R.M.†, Sánchez R., Rose, I., Hummer, G., Kühlbrandt, W., Vonck, J., Averhoff, B. (2017) Cryo-EM structure of the bifunctional secretin complex of Thermus thermophilus. eLife 6: e30483.
-
Ramachandran, R., Joseph, A.P., Bhaskara, R.M., Srinivasan, N. (2016) Structural and mechanistic insights into human splicing factor SF3b complex derived using an integrated approach guided by the cryo-EM density maps. RNA Biology 13 (10), 1025-1040
-
Craveur, P., Joseph, A.P., Esque, J., Narwani, T.J., Noël, F., Shinada, N., Goguet, M., Leonard, S., Poulain, P., Bertrand, O., Faure, G., Rebehmed, J., Ghozlane, A., Swapna, L.S., Bhaskara, R.M., Barnoud, J., Téletchéa, S., Jallu, V., Cerny, J., Schneider, B., Etchebest, C., Srinivasan, N., Gelly, J.G., de Brevern A.G. (2015) Protein flexibility in the light of structural alphabets. Front Mol. Biosci. 2 (20).
-
Bhaskara, R.M., Padhi, A., Srinivasan, N. (2014) Accurate prediction of interfacial residues in two-domain proteins using evolutionary information: implications for three-dimensional modeling. Proteins: Struct., Funct., Bioinf. 82 (7), 1219-1234.
-
Bhaskara, R.M., de Brevern, A. G., Srinivasan, N. (2012) Understanding the role of domain-domain linkers in the spatial orientation of domains in multi-domain proteins. J. Biomol. Struct. Dyn. 31(12), 1467-1480.
-
Swapna, L.S., Bhaskara, R.M., Sharma, J., Srinivasan, N. (2012) Roles of residues in the interface of transient protein-protein complexes before complexation. Sci. Rep. 2, 334.
We study biological pathways using various simulation methods. We focus on protein-induced membrane mechanics to reveal molecular mechanisms at cellular and organellar membranes (e.g., Autophagy pathways). Within these projects, we aim at capturing the dynamics of biomolecular systems at various temporal and spatial scales in quantitative terms to (i) provide mechanistic understanding & erect testable hypotheses, (ii) guide the design of new experiments, and (iii) provide a theoretical framework for experimental observations and simulations.
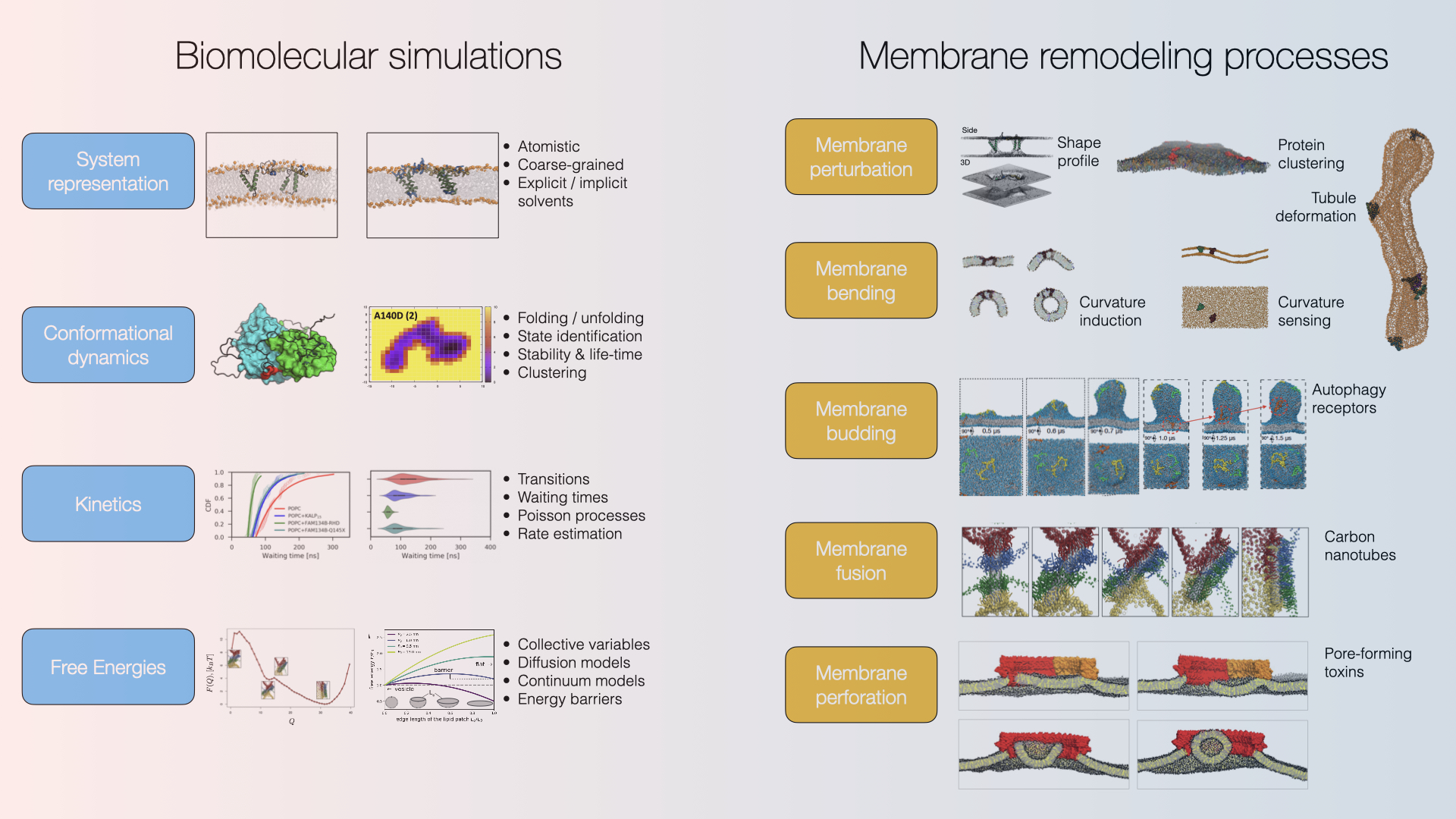
Selected publications
-
Reggio A., Bunomo, V., Berkane, R., Bhaskara, R. M., Tellechea, M., Peluso, I., Polischuk, E., Di Lorenzo, G., Crillo, C., Esposito, M., Hussain, A., Huebner, A.K., Settembre, C., Hummer, G., Grumati, P., Stolz, A. (2021) Role of FAM134 paralogues in endoplasmic reticulum remodeling, ER-phagy and Collagen quality control EMBO Rep. 22: e52289.
-
Ho, N. T., Siggel, M., Camacho, K. V., Bhaskara, R. M., Hicks, J. M., Yao, Y., Zhang, Y., Köfinger, J., Hummer, G., Noy, A. (2021) Membrane fusion and drug delivery with carbon nanotube porins. Proc. Natl. Acad. Sci. USA. 118, e2016974118.
-
Siggel, M., Bhaskara, R.M., Moesser, M.K., Dikic, I., Hummer, G. (2021) FAM134B-RHD protein clustering drives spontaneous budding of asymmetric membranes. J. Phys. Chem. Lett. 12 (7), 1926–1931.
-
Herhaus, L., Bhaskara, R.M.†, Lystad, A.H.†, Simonsen, A, Hummer, G., Dikic, I. (2020) TBK1-mediated phosphorylation of LC3C and GABARAPL2 controls autophagosome shedding by ATG4 protease. EMBO Rep. 21: e48317
-
Siggel, M., Bhaskara, R.M., Hummer, G. (2019) Phospholipid scramblases remodel the shape of asymmetric membranes. J. Phys. Chem. Lett. 10 (20), 6351-6354.
-
Vögele, M., Bhaskara, R.M., Mulvihill, E., van Pee, K., Yildiz, Ö., Kühlbrandt, W., Müller, D.J., Hummer, G. (2019) Membrane perforation by the pore-forming toxin pneumolysin. Proc. Natl. Acad. Sci. USA. 116 (27) 13352-13357.
-
Bhaskara, R.M., Grumati, P., Garcia-Padro, J., Kalayil, S., Covarrubais-Pinto., A., Chen, W., Kudrayshev, M., Dikic, I., Hummer, G. (2019) Curvature induction and membrane remodeling by FAM134B reticulon homology domain. Nat. Commun. 10:2370.
-
Bhaskara, R.M.†, Linker, S.M.†, Vöegele, M., Köfinger, J., Hummer, G. (2017) Carbon nanotubes mediate fusion of lipid vesicles. ACS Nano 11 (2), 1273-1280.
We employ data science approaches to integrate disparate datasets effectively. For example, we combine information from large-scale HTP datasets (Proteomic, Genetic, Chemical & Imaging) with bioinformatic predictions and publically available repositories to quantify data, mine and learn underlying biological patterns and uncover underlying processes.
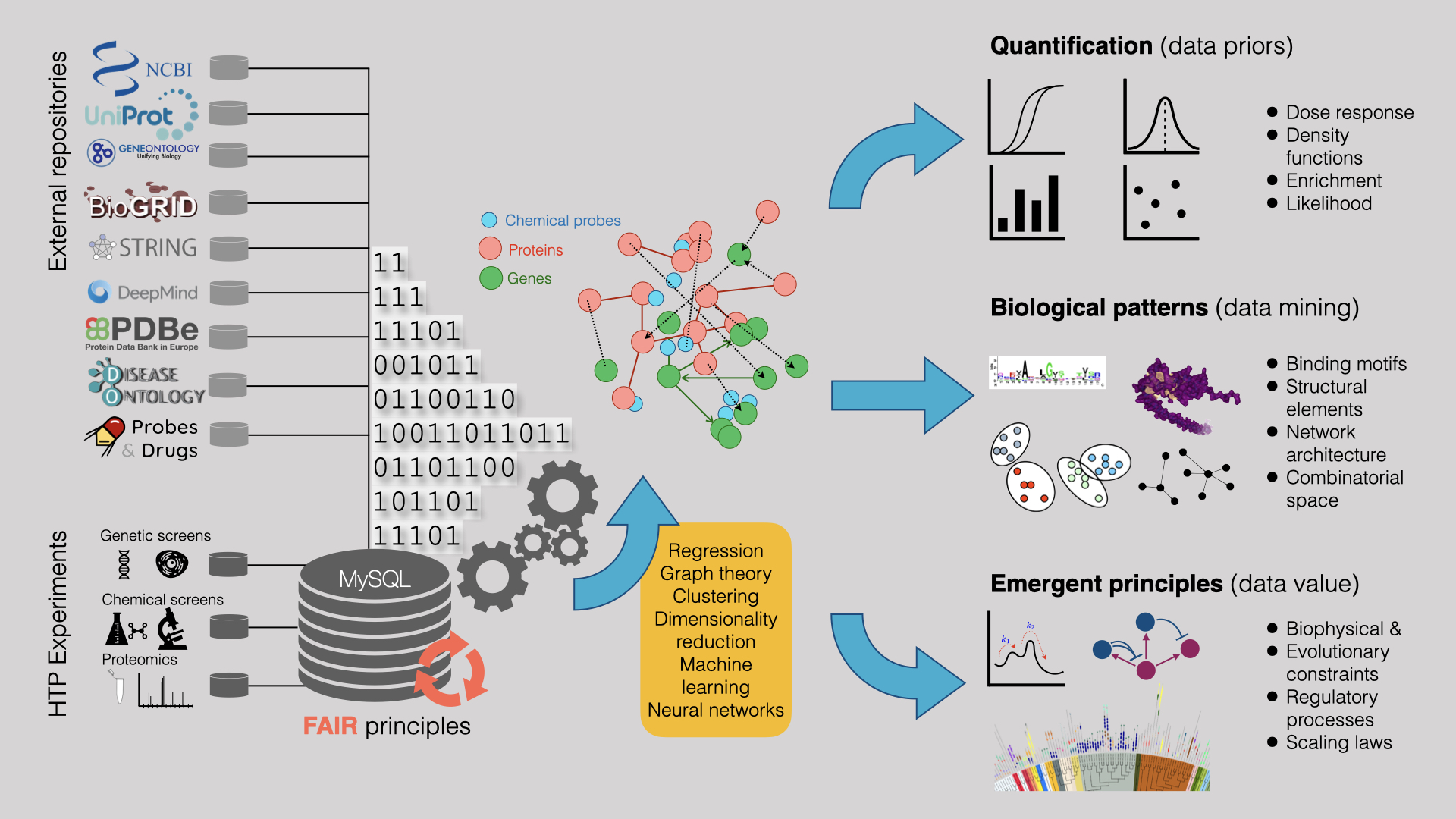
Selected publications
-
Mehrotra, P., Bhaskara, R.M., Gnanavel, M., Rakshambikai, R., Martin, J., Srinivasan, N. (2015) Classification of protein sequences with special references to multi-domain systems. J. Biomol. Struct. Dyn. 33 (sup1), 110
-
Gnanavel, M., Mehrotra, P., Rakshambikai, R., Martin, J., Srinivasan, N. Bhaskara, R.M. (2014) CLAP: A web-server for automatic classification of proteins with special reference to multi-domain proteins. BMC Bioinformatics.15 (343).
-
Bhaskara, R.M., Mehrotra, P., Rakshambikai, R., Gnanavel, M., Martin, J., Srinivasan, N. (2014) The relationship between classification of multidomain proteins using an alignment-free approach and their functions: a case study with immunoglobulins. Mol Biosyst. 10 (5), 1082-1093.
-
Bhaskara, R.M., and Srinivasan, N. (2011) Stability of domain structures in multi-domain proteins. Sci. Rep. 1, 40.
-
Srinivasan, N., Agarwal, G., Bhaskara, R.M., Gadkari, R., Krishnadev, O., Lakshmi, B., Mahajan, S., Mohanty, S., Mudgal, R., Rakshambikai, R., Sandhya, S., Sudha, G., Swapna, L.S., Tyagi, N. (2011) Influence of genomic and other biological data sets in the understanding of protein structures, functions, and interactions. International Journal of Knowledge Discovery in Bioinformatics (IJKDB) 2, 24-44.
Ramachandra M Bhaskara

PI
Email: bhaskara@med.uni-frankfurt.de
Fax: +49 69 798763-42581
Phone: +49 (0) 69 7984-2526
Ramachandra M Bhaskara

PI
Email: bhaskara@med.uni-frankfurt.de
Fax: +49 69 798763-42581
Phone: +49 (0) 69 7984-2526
All IBCII Members Contact Data

















