Gene Editing
Drug resistance
In cancer patient care, the National Cancer Institute (NCI) defines the term drug resistance as the failure of cancer cells to respond to a drug that used to kill or weaken them. Resistance to therapy can be divided into two broad categories: cell intrinsic and acquired mechanisms. Modern technologies such as deep-sequencing have aided in the interrogation of clinical samples and allowed the identification of molecular signatures and genotypes that predict response to a certain drug. From those data, a complex picture emerges, in which drug resistance can result from a wide range of molecular mechanisms: increased rates of drug efflux, alterations in drug metabolism and mutation of drug targets, as well as the activation of survival and inactivation of death pathways. Epigenetic changes as well as the cancer microenvironment have also been identified as important contributors to drug resistance. Hence, the molecular determinants of drug resistance can be complex and require innovative technologies to identify the underlying mechanisms.
CRISPR/Cas9
The CRISPR/Cas system has been described as a bacterial immune system, targeting viral pathogens such as lytic bacteriophages. The underlying mechanism is based on the fact that bacterial DNA nucleases can recognize and destroy bacteriophage DNA sequences by utilizing small guide RNAs through DNA double-strand cleavage. Interestingly, throughout this process, bacteria incorporate short sequences of the bacteriophage DNA into their own genomic DNA, thereby accumulating an encoded memory of pathogens that previously infected the bacteria. These incorporated short viral DNA sequences are arranged in form of clustered regularly-interspaced short palindromic repeats (CRISPR).
CRISPR/Cas gene editing depends on two components, a protein with DNA-nuclease activity (Cas) and a short RNA oligonucleotide (guide RNA, gRNA) that directs the nuclease activity of Cas enzymes to gRNA-complementary DNA sequences, thereby conferring specificity to the reaction. An additional prerequisite for successful DNA targeting of the Cas-gRNA complex is the presence of a protospacer-adjacent motif (PAM) DNA sequence in the target DNA, for which the exact sequence depends on the bacterial Cas-enzyme. For the most widely used Streptococcus pyogenes Cas9 (SpCas9), this sequence has the format of NGG, where N can be any nucleotide. Most notably, the Cas enzyme can be expressed in human cells and, by providing a human DNA-directed gRNA, induce a highly specific DNA double strand break that cannot be repaired error-free, leading to insertion and deletion (InDel) mutations. Phenotypes of InDel mutations range from in-frame deletions to complete gene knockouts.
While single genetic changes can be used to generate well-controlled model systems, they do not allow for unbiased screenings. To perform genetic screens, a multitude of gRNA sequences can be combined to generate libraries targeting open reading frames (ORFs) or else in the human or other genomes. Major advantages of these genetic screens are their unbiased performance and ease of use applicability to many biological questions.
gRNA library generation - limitations
State-of-the-art knockout gRNA library generation starts with the bioinformatical identification of functional gRNAs for which several online tools have become available (Table 1).
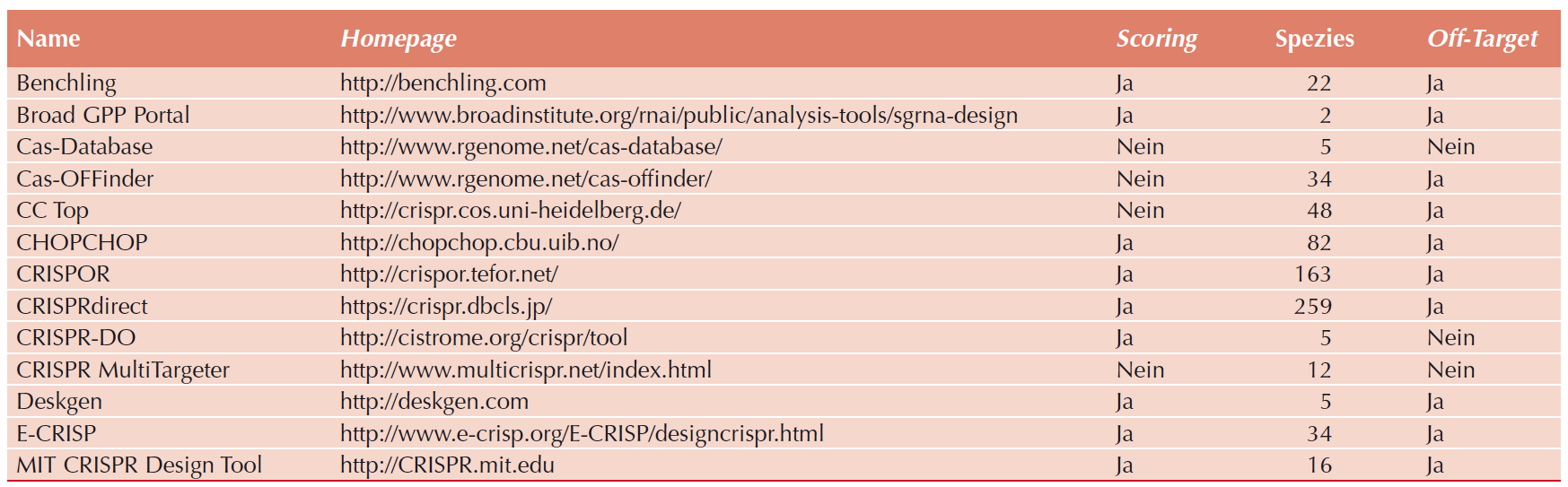 |
| Table 1: gRNA-design algorithms. Adapted from De Bruyn, et al., PHARMAKON, 2017. |
Identified sequences are extended by adapter sequences, coding for restriction enzyme recognition sites as well as primer annealing sites. Designed sequences are subsequently pooled synthesized, PCR amplified and enzymatically digested. Most commonly, the PCR product is introduced into lentiviral plasmids, requiring an open chromatin species of the target plasmid. Conventionally, gRNAs are pool-cloned by T4 DNA ligase or homology-based recombination procedures, resulting in the final gRNA library.
However, available gRNA libraries suffer from the methods they are generated with. Pooled gRNA cloning requires PCR amplification that introduces a PCR-dependent sequence bias of up to 10.000-fold into the library. This bias translates into high numbers of experimental coverage to ensure the experimental presence of all cloned sequences, making genome-wide screens unfeasible for most laboratories. Furthermore, restriction enzyme or homology-based cloning methods required open target-plasmid DNA. This open form of DNA is a substrate for the intrinsic exonuclease activity of most ligases, resulting in cloning artefacts and dramatically reducing final library quality.
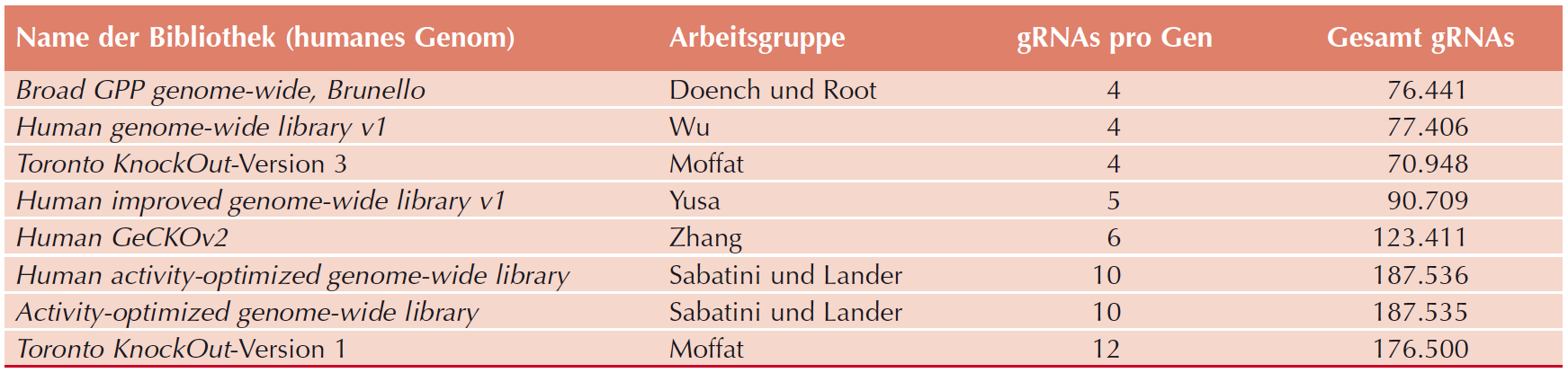 |
| Table 2: Available genome-wide gRNA-libraries. Adapted from De Bruyn, et al., PHARMAKON, 2017. |
Due to these technical issues, the maximal diversity of gRNA libraries is estimated to be between 100.000 and 300.000 gRNA sequences (Table 2). However, there are 250 million SpCas9 target sites in human genome, hence, conventional cloning methods cannot generate gRNA libraries with sufficient diversity to cover the coding as well as the non-coding human genome.
Therefore, functional genomic interrogations with the CRISPR/Cas system have been mostly focused on the coding genome.
Research in the Kaulich laboratory focusses on understanding malignant cell cycle entry and drug resistance mechanisms. To do so, we develop innovative technologies to functionally interrogate the human coding and non-coding genome.
Contact details
Dr. Manuel Kaulich
Institute of Biochemistry II
Goethe University Medical School
University Hospital Building 75
Theodor-Stern-Kai 7
60590 Frankfurt am Main / Germany
Office: +49 (0) 69 6301 5450
Lab: +49 (0) 69 6301
Email: kaulich (at) em.uni-frankfurt.de
Web: geg.biochem2.de
ResearchGate: https://www.researchgate.net/profile/Manuel_Kaulich
LinkedIn: https://www.linkedin.com/in/manuel-kaulich-ph-d-89705391/
Prof. Dr. Manuel Kaulich
Antonella Vera-Guapi
Antonella studied Genetics and Immunology at the University of Toronto in Canada. During her bachelor thesis, she studied the role of non-coding RNAs in cardiomyopathies. Afterwards, she moved to Germany to pursuit her Master’s degree in Molecular Medicine at the Friedrich Schiller University, Jena. For her MSc thesis she employed machine learning and statistics to identify host-dependency factors essential for the SARS-CoV-2 life cycle by using publicly available and own data from siRNA and drug screenings. In February 2022, Antonella joined the Gene Editing Group as a PhD student to investigate gene dependencies in chemo-resistant lung cancer.
Chiara Becht
Chiara studied molecular and technical medicine at Furtwangen University of applied Sciences, where she was interested in Biostatistics and Bioinformatics. Aiming for a deeper understanding of potential data analysis methods for the Life Sciences, Chiara joined the master program Data Science for Life Sciences at the Hanze University of applied Sciences. In her master thesis she focused on Knowledge Mining and Knowledge representation. Chiara joined the gene editing group in November 2023 as a PhD student to develop computational approaches for gene function inference from large-scale paired gene perturbations.
Dr. Cosmin Tudose
Cosmin graduated with an MSci Genetics degree from The University of Nottingham in 2020. Interested in bioinformatics and cancer biology, he did a PhD as part of the CRT Genomics Data Science at University College Dublin. His PhD work involved using biological networks to uncover vulnerabilities in childhood leukaemia. During this time he developed his expertise in omics data analysis (e.g., WGS, RNA-Seq, ATAC-Seq, CUT&RUN, Hi-C, CRISPR screens, etc.). He joined the Kaulich lab in March 2025 as an Alexander von Humboldt postdoctoral fellow. His work focuses on the analysis of genetic interaction data and computational approaches to integrate perturbation and omics data.

Dorothea Maneta
Coming from Athens, Greece, Dorothea obtained an Integrated Master’s diploma from the School of Applied Mathematical and Physical Sciences, National Technical University of Athens. For her thesis, she employed machine learning algorithms to predict cancer cell survival after exposure to ionizing radiation. Having developed an interest in biosciences, she enrolled in the MSc Molecular Biomedicine at the Medical School, University of Athens, and performed her thesis at BSRC Alexander Fleming. During her stay at the institute, she worked on the potential of gene editing in lung cancer animal models and the characterization of antigen-presenting cancer-associated fibroblasts in NSCLC patients. She joined the Gene Editing Group in February 2023 as a PhD student to investigate the modulation of ubiquitin pathways to enhance prime editing efficiencies.
Dr. Ilaria Persico
Ilaria was born in Trieste, Italy. She studied Functional Genomics at the University of Trieste and
obtained her MSc in 2019, focusing on the functional characterization of a splicing variant
responsible for Fanconi anemia (FA) disorder.
For her doctoral work, Ilaria received joint supervision from the University of Trieste and the Universitat Autònoma de Barcelona (Spain) where she
continued her research on FA diagnostic and therapeutic challenges. In this regard, she developed
an optimized mutation screening approach for patient’s exhaustive molecular characterization,
generated FA cellular models suitable for a repurposing drug screen, and dissected novel possible
synthetic interactions with FA deficiency by genome-wide CRISPR knockout screens. In 2023, she
joined the group of Manuel Kaulich with a prestigious Alexander von Humboldt postdoctoral
fellowship to establish a CRISPR-based prime-editing screening pipeline for the clinical
interpretation of pathogenic variants of unknown significance.
Miłosz Herka
Miłosz obtained his Bachelor’s degree in Biotechnology from Lodz University of Technology in Poland, where he developed a strong interest in genetic engineering. He subsequently completed a Master’s degree in Advanced Genetics at the Autonomous University of Barcelona (UAB), Spain. His master’s thesis focused on genetic interactions within the Fanconi anemia pathway, explored as potential therapeutic targets in translational and precision medicine. In 2024, he joined Prof. Manuel Kaulich’s laboratory, where his doctoral research centers on the interplay between transcriptional adaptation as a genetic buffering mechanism and nuclear speckles, as well as on the genetic dependencies in autophagy.
Privately, he's a committed activist and human rights educator, associated with Amnesty International and other local non-profit organisations.
Ronay Cetin
Ronay studied molecular biology, genetics and bioengineering at Sabanci University in Istanbul. In his Master’s Thesis Ronay used CRISPR/Cas9 editing to study transcription factors regulating IL7R gene expression in T lymphocytes. In January 2020, Ronay joined the Gene Editing Group of Dr. Manuel Kaulich as a PhD student to understand the role of cellular communication in oncogene-mediated transformation.
Sebastian Süsser
Sebastian completed a Bachelor’s degree in Biosciences and a Master’s degree in Molecular Medicine at Goethe University, including a one-year research stay at UCLA in Los Angeles as part of the Master’s program. During his Master’s thesis, he gained initial experience with CRISPR screening, which motivated him to pursue a PhD in this field. Sebastian joined the Kaulich lab at the end of 2022, where he is establishing combinatorial CRISPR screens at a genome-wide scale to uncover novel genetic dependencies and mechanisms of cellular gene regulation. His current research centers on understanding how cells orchestrate transcriptional responses to gene perturbations in order to achieve genetic robustness.
Simone Schaubeck
Dr. Yves Matthess
Yves received his diploma in Biology and Computer Science from the Goethe University of Frankfurt. He developed an inducible shRNA system and an shRNA prediction algorithm for RNA interference (RNAi) to uncover the functions of oncogenes. Applying the inducible RNAi system, a transgenic animal model was implemented to demonstrate cancer addiction of Polo-like kinase 1. Working on the cell cycle at the Institute of Gynecology and Obstetrics in Frankfurt he used RNAi screening technology and high-throughput data analysis for improving drug target opportunities for ovarian cancer patients. In 2018 he joined the IBC2 as postdoctoral scientist building up the FCSC platform. Recently he designed an inducible and multiplex-based CRISPR/Cas9 system for conditional gene knockouts.
Past Members:
Cover Shot’s
 |
 |
 |
 |
 |
HR-Info Radiointerview - CRISPR/Cas9
HR-Info Radiointerview von Manuel Kaulich zum Anlass des Paul Ehrlich und Ludwig Darmstädter-Preises 2016 an Jennifer Doudna und Emmanuelle Charpentier zum Thema CRISPR/Cas9 Technologie und dessen Anwendungen.
English:
HR-Info radio interview with Manuel Kaulich regarding the Paul Ehrlich and Ludwig Darmstädter Price 2016, awarded to Jennifer Doudna und Emmanuelle Charpentier for their contribution to the CRISPR/Cas9 gene-editing technology and its applications.
Dutta A, Cristiani A, Nikte SV, Eisert J, Matthess Y, Markusic B, Tudose C, Becht C, Shah VJ, Mosler T, Husnjak K, Dikic I, Kaulich M, Bhaskara RM Multi-scale classification decodes the complexity of the human E3 ligome. Nat Commun 2025. 16 (1) 11382 Link
Rohde T, Demirtas TY, Süsser S, Shaw AH, Kaulich M, Billmann M BaCoN (Balanced Correlation Network) improves prediction of gene buffering. Mol Syst Biol 2025. 21 (7) 807-824 Link
Prieto-Garcia C, Matkovic V, Mosler T, Li C, Liang J, Oo JA, Haidle F, Mačinković I, Cabrera-Orefice A, Berkane R, Giuliani G, Xu F, Jacomin AC, Tomaskovic I, Basoglu M, Hoffmann ME, Rathore R, Cetin R, Boutguetait D, Bozkurt S, Hernández Cañás MC, Keller M, Busam J, Shah VJ, Wittig I, et al. Pathogenic proteotoxicity of cryptic splicing is alleviated by ubiquitination and ER-phagy. Science 2024. 386 (6723) 768-776 Link
Kaulich M Long-noncoding vulnerabilities in MM. Blood 2024. 144 (16) 1654-1655 Link
Hsia O, Hinterndorfer M, Cowan AD, Iso K, Ishida T, Sundaramoorthy R, Nakasone MA, Imrichova H, Schätz C, Rukavina A, Husnjak K, Wegner M, Correa-Sáez A, Craigon C, Casement R, Maniaci C, Testa A, Kaulich M, Dikic I, Winter GE, Ciulli A Targeted protein degradation via intramolecular bivalent glues. Nature 2024. 627 (8002) 204-211 Link
Dönig J, Mende H, Davila Gallesio J, Wagner K, Hotz P, Schunck K, Piller T, Hölper S, Uhan S, Kaulich M, Wirth M, Keller U, Tascher G, Bohnsack KE, Müller S Characterization of nucleolar SUMO isopeptidases unveils a general p53-independent checkpoint of impaired ribosome biogenesis. Nat Commun 2023. 14 (1) 8121 Link
Sporbeck K, Haas ML, Pastor-Maldonado CJ, Schüssele DS, Hunter C, Takacs Z, Diogo de Oliveira AL, Franz-Wachtel M, Charsou C, Pfisterer SG, Gubas A, Haller PK, Knorr RL, Kaulich M, Macek B, Eskelinen EL, Simonsen A, Proikas-Cezanne T The ABL-MYC axis controls WIPI1-enhanced autophagy in lifespan extension. Commun Biol 2023. 6 (1) 872 Link
Wegner M, Kaulich M ReCo: automated NGS read-counting of single and combinatorial CRISPR gRNAs. Bioinformatics 2023. 39 (8) Link
Meyer LM, Koschade SE, Vischedyk JB, Thoelken M, Gubas A, Wegner M, Basoglu M, Knapp S, Kaulich M, Eimer S, Shaid S, Brandts CH Deciphering the mitophagy receptor network identifies a crucial role for OPTN (optineurin) in acute myeloid leukemia. Autophagy 2023. 19 (11) 2982-2996 Link
Cetin R, Wegner M, Luwisch L, Saud S, Achmedov T, Süsser S, Vera-Guapi A, Müller K, Matthess Y, Quandt E, Schaubeck S, Beisel CL, Kaulich M Optimized metrics for orthogonal combinatorial CRISPR screens. Sci Rep 2023. 13 (1) 7405 Link
Elcocks H, Brazel AJ, McCarron KR, Kaulich M, Husnjak K, Mortiboys H, Clague MJ, Urbé S FBXL4 ubiquitin ligase deficiency promotes mitophagy by elevating NIX levels. EMBO J 2023. 42 (13) e112799 Link
Hertel A, Alves LM, Dutz H, Tascher G, Bonn F, Kaulich M, Dikic I, Eimer S, Steinberg F, Bremm A USP32-regulated LAMTOR1 ubiquitination impacts mTORC1 activation and autophagy induction. Cell Rep 2022. 41 (10) 111653 Link
Michaelis JB, Brunstein ME, Bozkurt S, Alves L, Wegner M, Kaulich M, Pohl C, Münch C Protein import motor complex reacts to mitochondrial misfolding by reducing protein import and activating mitophagy. Nat Commun 2022. 13 (1) 5164 Link
Ong YT, Andrade J, Armbruster M, Shi C, Castro M, Costa ASH, Sugino T, Eelen G, Zimmermann B, Wilhelm K, Lim J, Watanabe S, Guenther S, Schneider A, Zanconato F, Kaulich M, Pan D, Braun T, Gerhardt H, Efeyan A, Carmeliet P, Piccolo S, Grosso AR, Potente M A YAP/TAZ-TEAD signalling module links endothelial nutrient acquisition to angiogenic growth. Nat Metab 2022. 4 (6) 672-682 Link
Eichler M, Distler U, Nasrullah U, Krishnan A, Kaulich M, Husnjak K, Eberhardt W, Rajalingam K, Tenzer S, Pfeilschifter J, Imre G The caspase-2 substrate p54nrb exhibits a multifaceted role in tumor cell death susceptibility via gene regulatory functions. Cell Death Dis 2022. 13 (4) 386 Link
Baker F, Polat IH, Abou-El-Ardat K, Alshamleh I, Thoelken M, Hymon D, Gubas A, Koschade SE, Vischedyk JB, Kaulich M, Schwalbe H, Shaid S, Brandts CH Metabolic Rewiring Is Essential for AML Cell Survival to Overcome Autophagy Inhibition by Loss of ATG3. Cancers (Basel) 2021. 13 (23) Link
Riegel K, Yurugi H, Schlöder J, Jonuleit H, Kaulich M, Kirschner F, Arnold-Schild D, Tenzer S, Schild H, Rajalingam K ERK5 modulates IL-6 secretion and contributes to tumor-induced immune suppression. Cell Death Dis 2021. 12 (11) 969 Link
Schmidt F, Marx A, Baumgarten N, Hebel M, Wegner M, Kaulich M, Leisegang MS, Brandes RP, Göke J, Vreeken J, Schulz MH Integrative analysis of epigenetics data identifies gene-specific regulatory elements. Nucleic Acids Res 2021. 49 (18) 10397-10418 Link
Kaulich M, Link VM, Lapek JD, Lee YJ, Glass CK, Gonzalez DJ, Dowdy SF A Cdk4/6-dependent phosphorylation gradient regulates the early to late G1 phase transition. Sci Rep 2021. 11 (1) 14736 Link
Zhang H, Cao X, Tang M, Zhong G, Si Y, Li H, Zhu F, Liao Q, Li L, Zhao J, Feng J, Li S, Wang C, Kaulich M, Wang F, Chen L, Li L, Xia Z, Liang T, Lu H, Feng XH, Zhao B A subcellular map of the human kinome. Elife 2021. 10 Link
Diehl V, Wegner M, Grumati P, Husnjak K, Schaubeck S, Gubas A, Shah VJ, Polat IH, Langschied F, Prieto-Garcia C, Müller K, Kalousi A, Ebersberger I, Brandts CH, Dikic I, Kaulich M Minimized combinatorial CRISPR screens identify genetic interactions in autophagy. Nucleic Acids Res 2021. 49 (10) 5684-5704 Link
Andrade J, Shi C, Costa ASH, Choi J, Kim J, Doddaballapur A, Sugino T, Ong YT, Castro M, Zimmermann B, Kaulich M, Guenther S, Wilhelm K, Kubota Y, Braun T, Koh GY, Grosso AR, Frezza C, Potente M Control of endothelial quiescence by FOXO-regulated metabolites. Nat Cell Biol 2021. 23 (4) 413-423 Link
Baumgarten N, Schmidt F, Wegner M, Hebel M, Kaulich M, Schulz MH Computational prediction of CRISPR-impaired non-coding regulatory regions. Biol Chem 2021. 402 (8) 973-982 Link
Cetin R, Quandt E, Kaulich M Functional Genomics Approaches to Elucidate Vulnerabilities of Intrinsic and Acquired Chemotherapy Resistance. Cells 2021. 10 (2) Link
Brandes RP, Dueck A, Engelhardt S, Kaulich M, Kupatt C, De Angelis MT, Leisegang MS, le Noble F, Moretti A, Müller OJ, Skryabin BV, Thum T, Wurst W DGK and DZHK position paper on genome editing: basic science applications and future perspective. Basic Res Cardiol 2021. 116 (1) 2 Link
Eck F, Phuyal S, Smith MD, Kaulich M, Wilkinson S, Farhan H, Behrends C ACSL3 is a novel GABARAPL2 interactor that links ufmylation and lipid droplet biogenesis. J Cell Sci 2020. 133 (18) Link
Keiten-Schmitz J, Wagner K, Piller T, Kaulich M, Alberti S, Müller S The Nuclear SUMO-Targeted Ubiquitin Quality Control Network Regulates the Dynamics of Cytoplasmic Stress Granules. Mol Cell 2020. 79 (1) 54-67.e7 Link
Trembinski DJ, Bink DI, Theodorou K, Sommer J, Fischer A, van Bergen A, Kuo CC, Costa IG, Schürmann C, Leisegang MS, Brandes RP, Alekseeva T, Brill B, Wietelmann A, Johnson CN, Spring-Connell A, Kaulich M, Werfel S, Engelhardt S, Hirt MN, Yorgan K, Eschenhagen T, Kirchhof L, Hofmann P, Jaé N, et al. Aging-regulated anti-apoptotic long non-coding RNA Sarrah augments recovery from acute myocardial infarction. Nat Commun 2020. 11 (1) 2039 Link
Bogucka K, Pompaiah M, Marini F, Binder H, Harms G, Kaulich M, Klein M, Michel C, Radsak MP, Rosigkeit S, Grimminger P, Schild H, Rajalingam K ERK3/MAPK6 controls IL-8 production and chemotaxis. Elife 2020. 9 Link
Wiechmann S, Maisonneuve P, Grebbin BM, Hoffmeister M, Kaulich M, Clevers H, Rajalingam K, Kurinov I, Farin HF, Sicheri F, Ernst A Conformation-specific inhibitors of activated Ras GTPases reveal limited Ras dependency of patient-derived cancer organoids. J Biol Chem 2020. 295 (14) 4526-4540 Link
Wegner M, Husnjak K, Kaulich M Unbiased and Tailored CRISPR/Cas gRNA Libraries by SynthesizingCovalently-closed-circular (3Cs) DNA. Bio Protoc 2020. 10 (1) e3472 Link
Lim R, Sugino T, Nolte H, Andrade J, Zimmermann B, Shi C, Doddaballapur A, Ong YT, Wilhelm K, Fasse JWD, Ernst A, Kaulich M, Husnjak K, Boettger T, Guenther S, Braun T, Krüger M, Benedito R, Dikic I, Potente M Deubiquitinase USP10 regulates Notch signaling in the endothelium. Science 2019. 364 (6436) 188-193 Link
Wegner M, Diehl V, Bittl V, de Bruyn R, Wiechmann S, Matthess Y, Hebel M, Hayes MG, Schaubeck S, Benner C, Heinz S, Bremm A, Dikic I, Ernst A, Kaulich M Circular synthesized CRISPR/Cas gRNAs for functional interrogations in the coding and noncoding genome. Elife 2019. 8 Link
Wegner MS, Schömel N, Gruber L, Örtel SB, Kjellberg MA, Mattjus P, Kurz J, Trautmann S, Peng B, Wegner M, Kaulich M, Ahrends R, Geisslinger G, Grösch S UDP-glucose ceramide glucosyltransferase activates AKT, promoted proliferation, and doxorubicin resistance in breast cancer cells. Cell Mol Life Sci 2018. 75 (18) 3393-3410 Link
Le Guerroué F, Eck F, Jung J, Starzetz T, Mittelbronn M, Kaulich M, Behrends C Autophagosomal Content Profiling Reveals an LC3C-Dependent Piecemeal Mitophagy Pathway. Mol Cell 2017. 68 (4) 786-796.e6 Link
Pitchai GP, Kaulich M, Bizard AH, Mesa P, Yao Q, Sarlos K, Streicher WW, Nigg EA, Montoya G, Hickson ID A novel TPR-BEN domain interaction mediates PICH-BEND3 association. Nucleic Acids Res 2017. 45 (19) 11413-11424 Link
Wesely J, Steiner M, Schnütgen F, Kaulich M, Rieger MA, Zörnig M Delayed Mesoderm and Erythroid Differentiation of Murine Embryonic Stem Cells in the Absence of the Transcriptional Regulator FUBP1. Stem Cells Int 2017. 2017 5762301 Link
van Wijk SJL, Fricke F, Herhaus L, Gupta J, Hötte K, Pampaloni F, Grumati P, Kaulich M, Sou YS, Komatsu M, Greten FR, Fulda S, Heilemann M, Dikic I Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-κB and restricts bacterial proliferation. Nat Microbiol 2017. 2 17066 Link
Lönn P, Kacsinta AD, Cui XS, Hamil AS, Kaulich M, Gogoi K, Dowdy SF Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics. Sci Rep 2016. 6 32301 Link
Meitinger F, Anzola JV, Kaulich M, Richardson A, Stender JD, Benner C, Glass CK, Dowdy SF, Desai A, Shiau AK, Oegema K 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J Cell Biol 2016. 214 (2) 155-66 Link
Tobias IS, Kaulich M, Kim PK, Simon N, Jacinto E, Dowdy SF, King CC, Newton AC Protein kinase Cζ exhibits constitutive phosphorylation and phosphatidylinositol-3,4,5-triphosphate-independent regulation. Biochem J 2016. 473 (4) 509-23 Link
Kaulich M, Dowdy SF Combining CRISPR/Cas9 and rAAV Templates for Efficient Gene Editing. Nucleic Acid Ther 2015. 25 (6) 287-96 Link
Kaulich M, Lee YJ, Lönn P, Springer AD, Meade BR, Dowdy SF Efficient CRISPR-rAAV engineering of endogenous genes to study protein function by allele-specific RNAi. Nucleic Acids Res 2015. 43 (7) e45 Link
Meade BR, Gogoi K, Hamil AS, Palm-Apergi C, van den Berg A, Hagopian JC, Springer AD, Eguchi A, Kacsinta AD, Dowdy CF, Presente A, Lönn P, Kaulich M, Yoshioka N, Gros E, Cui XS, Dowdy SF Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat Biotechnol 2014. 32 (12) 1256-61 Link
Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife 2014. 3 Link
Kaulich M, Cubizolles F, Nigg EA On the regulation, function, and localization of the DNA-dependent ATPase PICH. Chromosoma 2012. 121 (4) 395-408 Link
Fava LL, Kaulich M, Nigg EA, Santamaria A Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J 2011. 30 (16) 3322-36 Link
Hübner NC, Wang LH, Kaulich M, Descombes P, Poser I, Nigg EA Re-examination of siRNA specificity questions role of PICH and Tao1 in the spindle checkpoint and identifies Mad2 as a sensitive target for small RNAs. Chromosoma 2010. 119 (2) 149-65 Link
Hellenbroich Y, Kaulich M, Opitz S, Schwinger E, Zühlke C No association of the SCA1 (CAG)31 allele with Huntington's disease, myotonic dystrophy type 1 and spinocerebellar ataxia type 3. Psychiatr Genet 2004. 14 (2) 61-3 Link
Institute of Biochemistry II
University Hospital Frankfurt
Goethe University
Theodor-Stern-Kai 7 / Building 75
60590 Frankfurt am Main
Germany
Tel (office): +49 (0) 69 6301 5450
Tel (lab): +49 (0) 69 6301 6295
Fax: +49 (0) 69 6301 7603
All IBCII Members Contact Data















