Ubiquitin Signaling
Dynamic and reversible post-translational modifications by multifunctional protein ubiquitin regulate a variety of cellular processes including protein stability, DNA transcription and repair, endocytosis, cell cycle and division, apoptosis, autophagy, antigen processing, inflammation and immune response (1, 2). Research in Husnjak group is centred on the role of ubiquitin and ubiquitin-like proteins in protein degradation and regulation of proliferation, apoptosis and necroptosis. The group is especially interested in identifying novel players in linear ubiquitination, a type of atypical ubiquitination so far implicated in acquired and innate immunity, autoinflammation, lymphocyte development, genotoxic stress response and Parkinson´s disease, by utilizing multidisciplinary approaches ranging from biochemistry, molecular biology, cell culture, mass spectrometry, mouse models and collaboration-based structural studies.
Latest Research
The Author File: Koraljka Husnjak
References
Dr. Koraljka Husnjak
Koraljka was born in Croatia, where she studied molecular biology at the Faculty of Science, University of Zagreb. She received her Master of Science and PhD training at the Division of Molecular Medicine at the Rudjer Boskovic Institute headed by Kresimir Pavelic. Koraljka initially worked on oncogenic viruses and molecular mechanisms of cancer development. After joining the group of Ivan Dikic as a postdoctoral fellow, she shifted her focus on identification and characterization of novel ubiquitin- and ubiquitin-like binding domains by various approaches, including the identification of Rpn13 as a novel proteasomal ubiquitin receptor. She started her independent career in 2010 as a group leader at IBC2. Her group is interested in various aspects of atypical ubiquitination. In particular, her laboratory aims to decipher the functional roles of various ubiquitin-binding proteins in protein degradation, signal transduction and immune response. Her most recent work relates to the role of linear ubiquitination and novel linear ubiquitin-binding domains in TNFR1 signaling and cell death. Since January 2019 Koraljka is a team leader of the FCSC.
Dutta A, Cristiani A, Nikte SV, Eisert J, Matthess Y, Markusic B, Tudose C, Becht C, Shah VJ, Mosler T, Husnjak K, Dikic I, Kaulich M, Bhaskara RM Multi-scale classification decodes the complexity of the human E3 ligome. Nat Commun 2025. 16 (1) 11382 Link
Wagner K, Keiten-Schmitz J, Adhikari B, Patra U, Husnjak K, McNicoll F, Dormann D, Müller-McNicoll M, Tascher G, Wolf E, Müller S Induced proximity to PML protects TDP-43 from aggregation via SUMO-ubiquitin networks. Nat Chem Biol 2025. 21 (9) 1408-1419 Link
Herhaus L, Gestal-Mato U, Eapen VV, Mačinković I, Bailey HJ, Prieto-Garcia C, Misra M, Jacomin AC, Ammanath AV, Bagarić I, Michaelis J, Vollrath J, Bhaskara RM, Bündgen G, Covarrubias-Pinto A, Husnjak K, Zöller J, Gikandi A, Ribičić S, Bopp T, van der Heden van Noort GJ, Langer JD, Weigert A, Harper JW, Mancias JD, et al. IRGQ-mediated autophagy in MHC class I quality control promotes tumor immune evasion. Cell 2024. 187 (25) 7285-7302.e29 Link
Kliza KW, Song W, Pinzuti I, Schaubeck S, Kunzelmann S, Kuntin D, Fornili A, Pandini A, Hofmann K, Garnett JA, Stieglitz B, Husnjak K N4BP1 functions as a dimerization-dependent linear ubiquitin reader which regulates TNF signalling. Cell Death Discov 2024. 10 (1) 183 Link
Hsia O, Hinterndorfer M, Cowan AD, Iso K, Ishida T, Sundaramoorthy R, Nakasone MA, Imrichova H, Schätz C, Rukavina A, Husnjak K, Wegner M, Correa-Sáez A, Craigon C, Casement R, Maniaci C, Testa A, Kaulich M, Dikic I, Winter GE, Ciulli A Targeted protein degradation via intramolecular bivalent glues. Nature 2024. 627 (8002) 204-211 Link
Berkane R, Ho-Xuan H, Glogger M, Sanz-Martinez P, Brunello L, Glaesner T, Kuncha SK, Holzhüter K, Cano-Franco S, Buonomo V, Cabrerizo-Poveda P, Balakrishnan A, Tascher G, Husnjak K, Juretschke T, Misra M, González A, Dötsch V, Grumati P, Heilemann M, Stolz A The function of ER-phagy receptors is regulated through phosphorylation-dependent ubiquitination pathways. Nat Commun 2023. 14 (1) 8364 Link
Brat C, Huynh Phuoc HP, Awad O, Parmar BS, Hellmuth N, Heinicke U, Amr S, Grimmer J, Sürün D, Husnjak K, Carlsson M, Fahrer J, Bauer T, Krieg SC, Manolikakes G, Zacharowski K, Steinhilber D, Münch C, Maier TJ, Roos J Endogenous anti-tumorigenic nitro-fatty acids inhibit the ubiquitin-proteasome system by directly targeting the 26S proteasome. Cell Chem Biol 2023. 30 (10) 1277-1294.e12 Link
Elcocks H, Brazel AJ, McCarron KR, Kaulich M, Husnjak K, Mortiboys H, Clague MJ, Urbé S FBXL4 ubiquitin ligase deficiency promotes mitophagy by elevating NIX levels. EMBO J 2023. 42 (13) e112799 Link
Eichler M, Distler U, Nasrullah U, Krishnan A, Kaulich M, Husnjak K, Eberhardt W, Rajalingam K, Tenzer S, Pfeilschifter J, Imre G The caspase-2 substrate p54nrb exhibits a multifaceted role in tumor cell death susceptibility via gene regulatory functions. Cell Death Dis 2022. 13 (4) 386 Link
Diehl V, Wegner M, Grumati P, Husnjak K, Schaubeck S, Gubas A, Shah VJ, Polat IH, Langschied F, Prieto-Garcia C, Müller K, Kalousi A, Ebersberger I, Brandts CH, Dikic I, Kaulich M Minimized combinatorial CRISPR screens identify genetic interactions in autophagy. Nucleic Acids Res 2021. 49 (10) 5684-5704 Link
Kliza K, Husnjak K Resolving the Complexity of Ubiquitin Networks. Front Mol Biosci 2020. 7 21 Link
Wegner M, Husnjak K, Kaulich M Unbiased and Tailored CRISPR/Cas gRNA Libraries by SynthesizingCovalently-closed-circular (3Cs) DNA. Bio Protoc 2020. 10 (1) e3472 Link
Lim R, Sugino T, Nolte H, Andrade J, Zimmermann B, Shi C, Doddaballapur A, Ong YT, Wilhelm K, Fasse JWD, Ernst A, Kaulich M, Husnjak K, Boettger T, Guenther S, Braun T, Krüger M, Benedito R, Dikic I, Potente M Deubiquitinase USP10 regulates Notch signaling in the endothelium. Science 2019. 364 (6436) 188-193 Link
Kniss A, Schuetz D, Kazemi S, Pluska L, Spindler PE, Rogov VV, Husnjak K, Dikic I, Güntert P, Sommer T, Prisner TF, Dötsch V Chain Assembly and Disassembly Processes Differently Affect the Conformational Space of Ubiquitin Chains. Structure 2018. 26 (2) 249-258.e4 Link
Kliza K, Taumer C, Pinzuti I, Franz-Wachtel M, Kunzelmann S, Stieglitz B, Macek B, Husnjak K Internally tagged ubiquitin: a tool to identify linear polyubiquitin-modified proteins by mass spectrometry. Nat Methods 2017. 14 (5) 504-512 Link
Husnjak K, Keiten-Schmitz J, Müller S Identification and Characterization of SUMO-SIM Interactions. Methods Mol Biol 2016. 1475 79-98 Link
Leznicki P, Korac-Prlic J, Kliza K, Husnjak K, Nyathi Y, Dikic I, High S Binding of SGTA to Rpn13 selectively modulates protein quality control. J Cell Sci 2015. 128 (17) 3187-96 Link
Tomasovic A, Kurrle N, Sürün D, Heidler J, Husnjak K, Poser I, Schnütgen F, Scheibe S, Seimetz M, Jaksch P, Hyman A, Weissmann N, von Melchner H Sestrin 2 protein regulates platelet-derived growth factor receptor β (Pdgfrβ) expression by modulating proteasomal and Nrf2 transcription factor functions. J Biol Chem 2015. 290 (15) 9738-52 Link
Aguileta MA, Korac J, Durcan TM, Trempe JF, Haber M, Gehring K, Elsasser S, Waidmann O, Fon EA, Husnjak K The E3 ubiquitin ligase parkin is recruited to the 26 S proteasome via the proteasomal ubiquitin receptor Rpn13. J Biol Chem 2015. 290 (12) 7492-505 Link
Husnjak K, Dikic I Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem 2012. 81 291-322 Link
Grabbe C, Husnjak K, Dikic I The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol 2011. 12 (5) 295-307 Link
Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 2008. 453 (7194) 548-52 Link
Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008. 453 (7194) 481-8 Link
Husnjak K, Dikic I EGFR trafficking: parkin' in a jam. Nat Cell Biol 2006. 8 (8) 787-8 Link
Schmidt MHH, Husnjak K, Szymkiewicz I, Haglund K, Dikic I Cbl escapes Cdc42-mediated inhibition by downregulation of the adaptor molecule betaPix. Oncogene 2006. 25 (21) 3071-8 Link
Sirotkovic-Skerlev M, Krizanac S, Kapitanovic S, Husnjak K, Unusic J, Pavelic K Expression of c-myc, erbB-2, p53 and nm23-H1 gene product in benign and malignant breast lesions: coexpression and correlation with clinicopathologic parameters. Exp Mol Pathol 2005. 79 (1) 42-50 Link
Kowanetz K, Husnjak K, Höller D, Kowanetz M, Soubeyran P, Hirsch D, Schmidt MHH, Pavelic K, De Camilli P, Randazzo PA, Dikic I CIN85 associates with multiple effectors controlling intracellular trafficking of epidermal growth factor receptors. Mol Biol Cell 2004. 15 (7) 3155-66 Link
Matovina M, Husnjak K, Milutin N, Ciglar S, Grce M Possible role of bacterial and viral infections in miscarriages. Fertil Steril 2004. 81 (3) 662-9 Link
Grce M, Husnjak K, Matovina M, Milutin N, Magdic L, Husnjak O, Pavelic K Human papillomavirus, cytomegalovirus, and adeno-associated virus infections in pregnant and nonpregnant women with cervical intraepithelial neoplasia. J Clin Microbiol 2004. 42 (3) 1341-4 Link
Grce M, Husnjak K, Milutin N, Matovina M [Detection of human papillomaviruses and other agents causing sexually transmitted diseases with molecular diagnosis methods]. Acta Med Croatica 2003. 57 (4) 295-301 Link
Kowanetz K, Szymkiewicz I, Haglund K, Kowanetz M, Husnjak K, Taylor JD, Soubeyran P, Engstrom U, Ladbury JE, Dikic I Identification of a novel proline-arginine motif involved in CIN85-dependent clustering of Cbl and down-regulation of epidermal growth factor receptors. J Biol Chem 2003. 278 (41) 39735-46 Link
Kralj M, Husnjak K, Körbler T, Pavelić J Endogenous p21WAF1/CIP1 status predicts the response of human tumor cells to wild-type p53 and p21WAF1/CIP1 overexpression. Cancer Gene Ther 2003. 10 (6) 457-67 Link
Skerlev M, Grce M, Sirotkoviae-Skerlev M, Husnjak K, Lipozencić J Human papillomavirus male genital infections: clinical variations and the significance of DNA typing. Clin Dermatol 2002. 20 (2) 173-8 Link
Grce M, Husnjak K, Bozikov J, Magdić L, Zlacki M, Lukac J, Fistonić I, Sikanić-Dugic N, Pavelić K Evaluation of genital human papillomavirus infections by polymerase chain reaction among Croatian women. Anticancer Res 2001. 21 (1B) 579-84 Link
Husnjak K, Grce M, Magdić L, Pavelić K Comparison of five different polymerase chain reaction methods for detection of human papillomavirus in cervical cell specimens. J Virol Methods 2000. 88 (2) 125-34 Link
Grce M, Husnjak K, Skerlev M, Lipozencić J, Pavelić K Detection and typing of human papillomaviruses by means of polymerase chain reaction and fragment length polymorphism in male genital lesions. Anticancer Res 2000. 20 (3B) 2097-102 Link
Grce M, Husnjak K, Magdić L, Ilijas M, Zlacki M, Lepusić D, Lukac J, Hodek B, Grizelj V, Kurjak A, Kusić Z, Pavelić K Detection and typing of human papillomaviruses by polymerase chain reaction in cervical scrapes of Croatian women with abnormal cytology. Eur J Epidemiol 1997. 13 (6) 645-51 Link
Grce M, Furcić I, Hrasćan R, Husnjak K, Krhen I, Mareković Z, Zeljko Z, Pavelić K Human papillomaviruses are not associated with renal carcinoma. Anticancer Res 1997. 17 (3C) 2193-6 Link
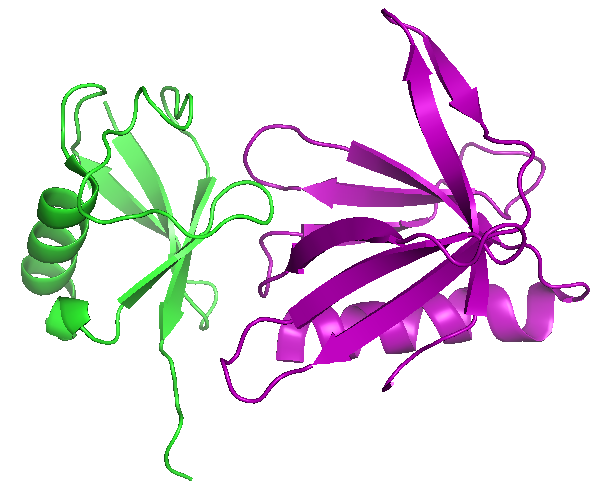
We identified and characterized Rpn13 as a proteasomal ubiquitin receptor (Figure 1) and determined its cellular function and structure of its ubiquitin-binding domain (UBD) Pru on the collaborative basis (1, 2). For the last several years the laboratory is working on the knock-out and ubiquitin-binding deficient knock-in models of Rpn13. Additionally, we could show how E3 ligase Parkin, relevant for the development of Parkinson disease, gets recruited and activated at the 19S proteasome through the interaction with Rpn13. We demonstrated that turnover of Parkin and several of its substrates involved in mitophagy, are impaired in the absence of Rpn13 (3). On the collaborative basis, we recently showed how Sestrin 2 protein acts as a positive regulator of proteasomal function and activates transcription of Nrf2-regulated antioxidant genes (4). We could also show that binding of SGTA (cytosolic factor implicated in the quality control of mislocalized membrane proteins) to Rpn13 selectively modulates protein quality control (5).
 |
|
Figure 1. Complex structures of mouse Rpn13 (22-130) and ubiquitin (PDB entry 2z59) |
References
1. Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters K, Finley D, Dikic I (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008; 453 (7194): 481-488.
2. Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters K, Groll M. Ubiquitin docking at the proteasome via a novel PH domain interaction. Nature 2008; 453 (7194): 548-552.
3. Aguileta MA, Korac J, Durcan TM, Trempe JF, Haber M, Gehring K, Elsasser S, Waidmann O, Fon EA, Husnjak K. The E3 Ubiquitin ligase parkin is recruited to the 26S proteasome via the proteasomal ubiquitin receptor Rpn13. J Biol Chem 2015; 290 (12): 7492-7505.
4. Tomasovic A, Kurrle N, Sürün D, Heidler J, Husnjak K, Poser I, Schnütgen F, Scheibe S, Seimetz M, Jaksch P, Hyman A, Weissmann N, von Melchner H. Sestrin 2 protein regulates platelet-derived growth factor receptor β (Pdgfrβ) expression by modulating proteasomal and Nrf2 transcription factor functions. J Biol Chem 2015; 290(15): 9738-9752.
5. Leznicki P, Prlic JK, Kliza K, Husnjak K, Nyathi Y, Dikic I, High S. SGTA binding to Rpn13 selectively modulates protein quality control. J Cell Sci 2015; 128(17): 3187–3196.
The most recently discovered modification by ubiquitin is linear polyubiquitination, which (unlike lysine-specific ubiquitination) relies on the formation of the peptide bond between the N-terminal methionine of one ubiquitin and the terminal glycine of another ubiquitin moiety. Even though only a single E3 ligase complex (LUBAC), one highly specific deubiquitinase (OTULIN) and a small number of substrates are currently known for linear polyubiquitination, multiple studies confirmed the crucial role of linear polyubiquitination in innate and adaptive immune signaling. The number of identified linear polyubiquitin-modified targets is rather small due to technical and experimental limitations.
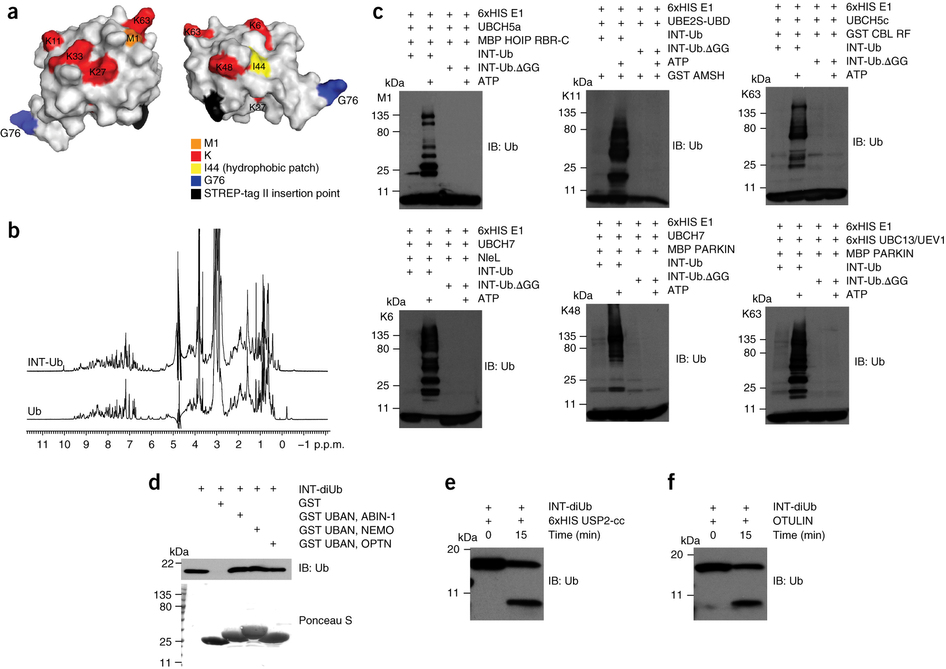
Our aim is to identify novel players in linear polyubiquitination, with emphasis on novel linear polyubiquitin-modified substrates. We therefore developed, validated and applied lysine-less internally tagged ubiquitin (INT-Ub.7KR) and combined it with mass spectrometry, in order to discover linear polyubiquitin-modified substrates, as well as novel processes where these proteins play a role (1).
 |
|
Figure 1. The design and in vitro validation of internally tagged ubiquitin (INT-Ub) (from Kliza et al, 2017) |
Reference
Polyubiquitin chains modifying target proteins are recognized by more than 20 families of ubiquitin receptors, i.e. proteins containing ubiquitin-binding domains (UBDs), which decode ubiquitinated target signals into biochemical cascades within the cell (1).
A very few linear polyubiquitin-specific UBDs have been described so far. In order to further expand the functional analysis of linear ubiquitin receptors that recognize and propagate linear ubiquitin signalling in the cells, my laboratory has performed yeast two-hybrid assays with tandem repeats of ubiquitin to identify novel LUBIDs and we are currently studying the physiological functions of identified LUBID- containing proteins.
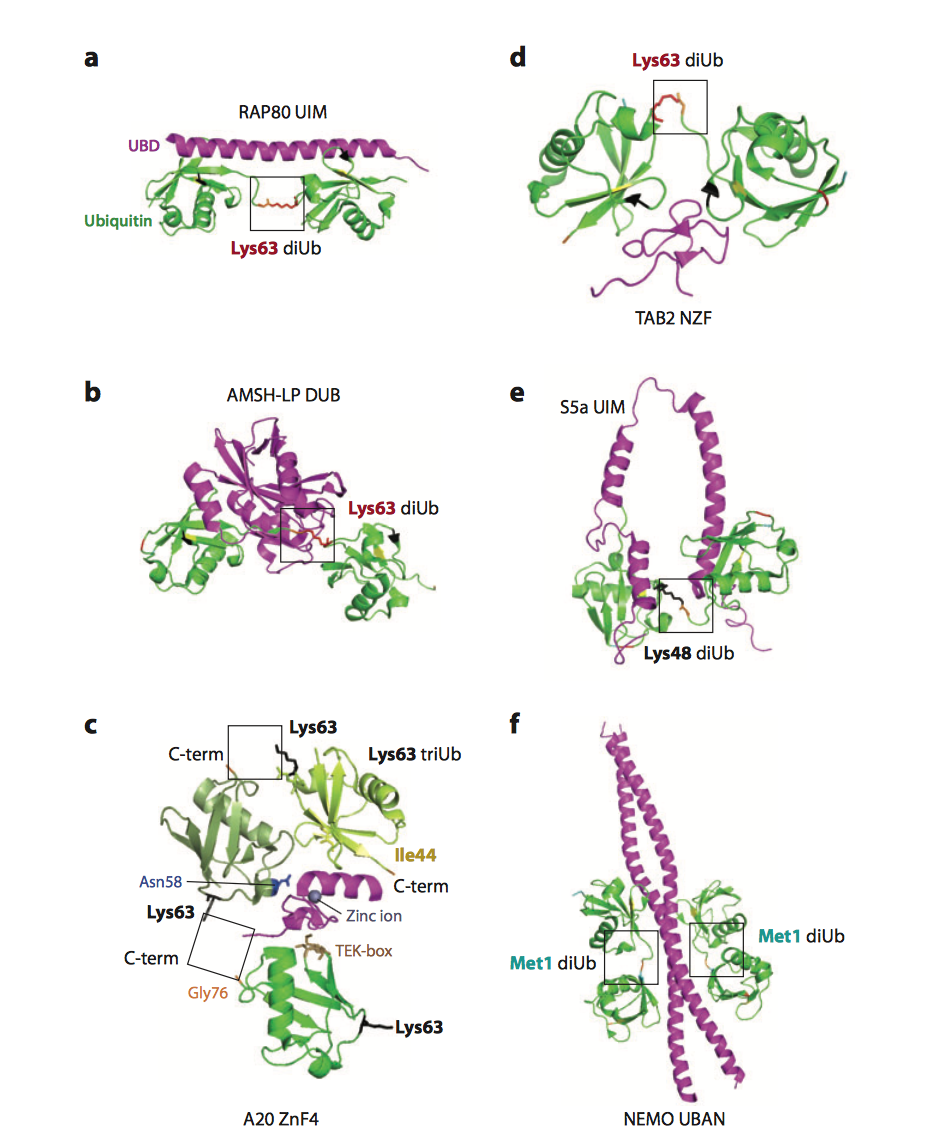 |
| Figure 1. Various ubiquitin receptors specifically recognize various ubiquitin linkages (from Husnjak and Dikic, 2012) |
