Molecular Cell Biology
Protein ubiquitination emerged as a fundamental regulatory process in the cell, and both proteolytic and non-proteolytic functions have been attributed to this small protein modifier. The various cellular responses to ubiquitin signals are highly regulated by factors that recognize or remove the modification (Ubiquitin-binding domain (UBD)-containing proteins and deubiquitinating enzymes (DUBs), respectively). Ubiquitin is attached to substrate lysine residues and can assemble at least eight differently linked polymers. Importantly, these ubiquitin chains have been associated with distinct cellular outputs, further increasing the astonishing complexity in the ubiquitin system.
Our lab investigates the role of protein ubiquitination in adaptive responses to cellular stress, e.g. hypoxia, oxidative damage or starvation. In this context, we are particularly interested in the function and regulation of DUBs. Altered DUB activity is associated with a multitude of pathologies including cancer and Alzheimer’s diseases. Therefore, DUBs represent promising novel structures for targeted therapy. We aim to increase our understanding of the physiological functions and control mechanisms of these important and versatile enzymes.
 |
|
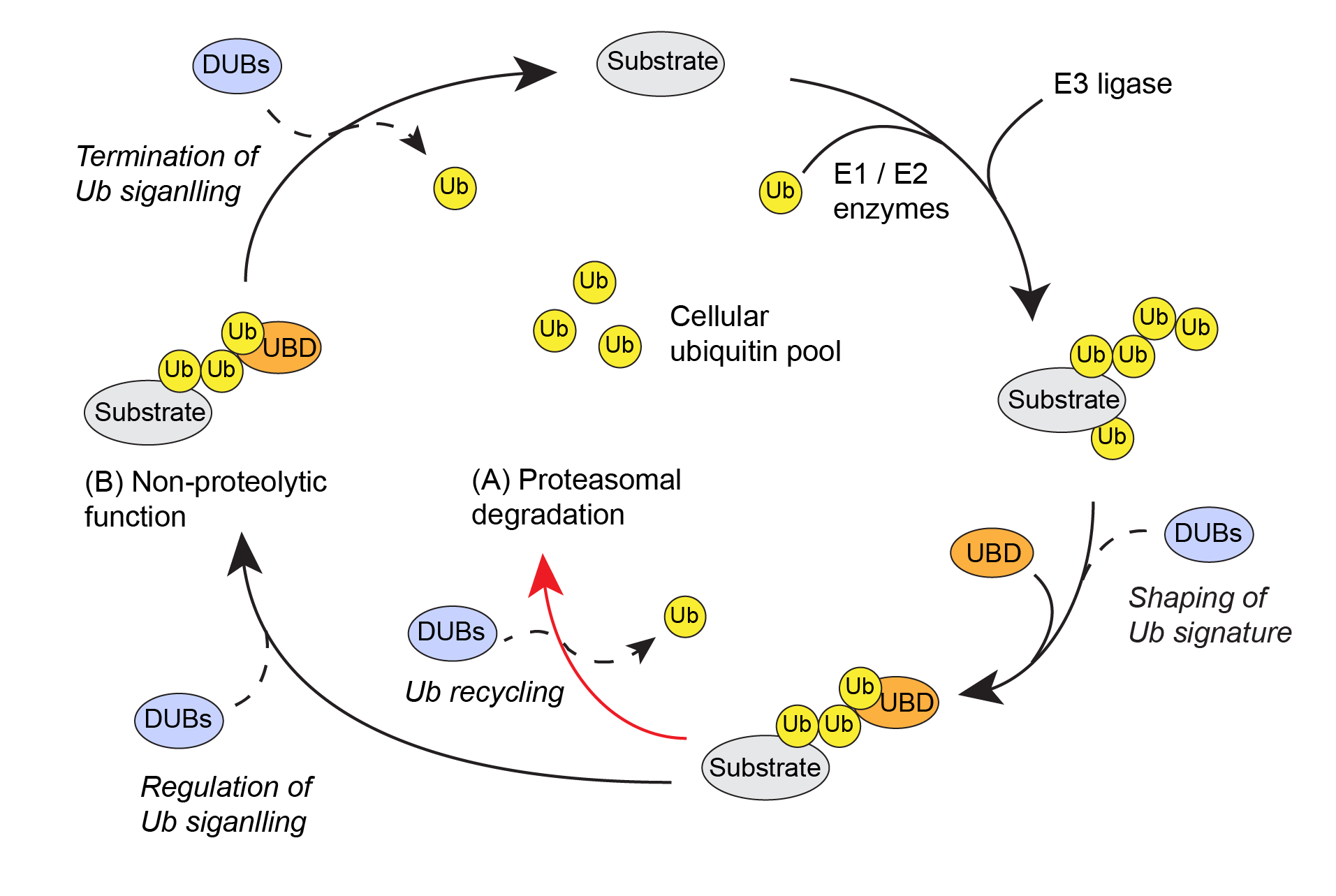
Figure 1: Schematic overview of the ubiquitin system. DUBs play essential roles in the correct outcome of the ubiquitin code. |
Dr. Anja Bremm
Alexandra Hertel
Justyna Bajdzienko
Justyna comes from Poland. She obtained her Bachelor’s degree in Biotechnology at the University of Wroclaw, where she studied expression profiles of selected genes of the biosynthetic gene cluster of the novel aminocoumarins cacibiocin A and B. She continued her Master’s studies in the group of Prof. Jolanta Zakrzewska-Czerwinska at the Department of Molecular Microbiology, University of Wroclaw. During her Master thesis she identified and studied a new protein involved in the regulation of replication initiation and differentiation in Streptomyces coelicolor. After graduation, Justyna moved to Germany and started an internship in Prof. Lothar Willmitzer’s group at the Max Planck Institute for Molecular Plant Physiology in Potsdam. She focused on validating the interaction partners of pantothenic acid and methylthioadenosine using microscale thermophoresis and thermo stability assays. Justyna also gained substantial experience in working with mass spectrometry-based metabolomics, which she made use of when she moved to China in 2018, where she participated in setting up a new mass spectrometry unit at Metanotitia, a Chinese metabolomics company in Shenzhen. In July 2020, Justyna returned to academia and joined the Bremm laboratory as PhD student. She is now investigating the functional role of deubiquitylases in selective autophagy pathways.
Ramya Lakshmana Iyer
Originally from Bangalore, India, Ramya obtained a Bachelor of Engineering degree in Biotechnology from M.S. Ramaiah Institute of Technology, India. She moved to Germany in 2017 to pursue Master of Science in Molecular Bioscience major in Cancer Biology from Ruprecht-Karls Universität Heidelberg. As a part of her master's thesis, she focused on studying the Role of ID3 in DNA repair at the Division of Cancer Epigenomics headed by Dr. Christoph Plass at the German Cancer Research Center. After graduation, Ramya joined the Team of DNA damage in Cancer at BioMed X GmbH, Heidelberg, where she worked on Pooled CRISPR screens in RPE1 cells modeling non-small cell lung cancer (NSCLC). She started her Ph.D. in Bremm lab in June 2021 to study the aberrant Nrf2 activity in NSCLC.
Past Members:
Fitzgerald JC, Bremm A A structural switch reveals how to disarm USP30 and unlock mitophagy. Nat Struct Mol Biol 2025. 32 (9) 1586-1588 Link
Bajdzienko J, Bremm A Mammalian pexophagy at a glance. J Cell Sci 2024. 137 (9) Link
Mende H, Khatri A, Lange C, Poveda-Cuevas SA, Tascher G, Covarrubias-Pinto A, Löhr F, Koschade SE, Dikic I, Münch C, Bremm A, Brunetti L, Brandts CH, Uckelmann H, Dötsch V, Rogov VV, Bhaskara RM, Müller S An atypical GABARAP binding module drives the pro-autophagic potential of the AML-associated NPM1c variant. Cell Rep 2023. 42 (12) 113484 Link
Hertel A, Eimer S, Bremm A LAMTOR1 ubiquitination restricts its interaction with the vacuolar-type H-ATPase, promotes autophagy and is controlled by USP32. Autophagy 2023. 19 (8) 2406-2407 Link
de Oliveira Freitas Machado C, Schafranek M, Brüggemann M, Hernández Cañás MC, Keller M, Di Liddo A, Brezski A, Blümel N, Arnold B, Bremm A, Wittig I, Jaé N, McNicoll F, Dimmeler S, Zarnack K, Müller-McNicoll M Poison cassette exon splicing of SRSF6 regulates nuclear speckle dispersal and the response to hypoxia. Nucleic Acids Res 2023. 51 (2) 870-890 Link
Hertel A, Alves LM, Dutz H, Tascher G, Bonn F, Kaulich M, Dikic I, Eimer S, Steinberg F, Bremm A USP32-regulated LAMTOR1 ubiquitination impacts mTORC1 activation and autophagy induction. Cell Rep 2022. 41 (10) 111653 Link
Bremm A Hug and hold tight: Dimerization controls the turnover of the ubiquitin-conjugating enzyme UBE2S. Sci Signal 2020. 13 (654) Link
Mader J, Huber J, Bonn F, Dötsch V, Rogov VV, Bremm A Oxygen-dependent asparagine hydroxylation of the ubiquitin-associated (UBA) domain in Cezanne regulates ubiquitin binding. J Biol Chem 2020. 295 (8) 2160-2174 Link
Wegner M, Diehl V, Bittl V, de Bruyn R, Wiechmann S, Matthess Y, Hebel M, Hayes MG, Schaubeck S, Benner C, Heinz S, Bremm A, Dikic I, Ernst A, Kaulich M Circular synthesized CRISPR/Cas gRNAs for functional interrogations in the coding and noncoding genome. Elife 2019. 8 Link
Fottner M, Brunner AD, Bittl V, Horn-Ghetko D, Jussupow A, Kaila VRI, Bremm A, Lang K Site-specific ubiquitylation and SUMOylation using genetic-code expansion and sortase. Nat Chem Biol 2019. 15 (3) 276-284 Link
Akutsu M, Dikic I, Bremm A Ubiquitin chain diversity at a glance. J Cell Sci 2016. 129 (5) 875-80 Link
Moniz S, Bandarra D, Biddlestone J, Campbell KJ, Komander D, Bremm A, Rocha S Cezanne regulates E2F1-dependent HIF2α expression. J Cell Sci 2015. 128 (16) 3082-93 Link
Bremm A, Moniz S, Mader J, Rocha S, Komander D Cezanne (OTUD7B) regulates HIF-1α homeostasis in a proteasome-independent manner. EMBO Rep 2014. 15 (12) 1268-77 Link
Dikic I, Bremm A DUBs counteract parkin for efficient mitophagy. EMBO J 2014. 33 (21) 2442-3 Link
Bremm A, Komander D Synthesis and analysis of K11-linked ubiquitin chains. Methods Mol Biol 2012. 832 219-28 Link
Bremm A, Komander D Emerging roles for Lys11-linked polyubiquitin in cellular regulation. Trends Biochem Sci 2011. 36 (7) 355-63 Link
Bremm A, Komander D A further case of Dop-ing in bacterial pupylation. EMBO Rep 2010. 11 (10) 722-3 Link
Bremm A, Freund SM, Komander D Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol 2010. 17 (8) 939-47 Link
Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol 2009. 16 (12) 1328-30 Link
Markson G, Kiel C, Hyde R, Brown S, Charalabous P, Bremm A, Semple J, Woodsmith J, Duley S, Salehi-Ashtiani K, Vidal M, Komander D, Serrano L, Lehner P, Sanderson CM Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res 2009. 19 (10) 1905-11 Link
Bremm A, Walch A, Fuchs M, Mages J, Duyster J, Keller G, Hermannstädter C, Becker KF, Rauser S, Langer R, von Weyhern CH, Höfler H, Luber B Enhanced activation of epidermal growth factor receptor caused by tumor-derived E-cadherin mutations. Cancer Res 2008. 68 (3) 707-14 Link
Ubiquitination plays an essential role in modulating protein functions and deregulation of the ubiquitin system leads to the development of multiple human diseases. Ubiquitin is covalently attached via its C-terminus to substrate lysine residues by a well-orchestrated enzymatic cascade including an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligases. Ubiquitin itself contains seven lysine residues and an N-terminal amino group, all of which can be linked to another ubiquitin to form polymers of eight different linkage types. It was shown that differently linked ubiquitin chains trigger distinct cellular responses, suggesting that ubiquitin can act as a code to store and transmit information.
Given the large complexity of possible ubiquitin modifications, the appropriate removal of ubiquitin by DUBs presents a significant problem to the cell. DUBs are essential to maintain free ubiquitin levels, rescue proteins from ubiquitin-mediated degradation, and control the dynamics of ubiquitin-mediated signaling events. Thus, the activity of DUBs must be tightly regulated in order to recognize both the correct substrate and the correct context in which to deubiquitinate target proteins. In addition to their physiological function, we are investigating cellular mechanisms of how the catalytic activity of these proteases is controlled (e.g. by PTMs, protein binding partners, or reactive oxygen species (ROS)), especially in adaptive cellular responses to hypoxia and oxidative stress.
Autophagy selectively delivers cytoplasmic components to lysosomes for degradation and recycling. Increasing evidence suggests that protein ubiquitination is directly involved in the regulation of autophagy by (a) controlling stability of upstream regulators or components of the autophagy machinery, and (b) facilitating the recruitment of autophagy adaptors. While a variety of E3 ubiquitin ligases were already demonstrated to govern autophagic substrate degradation, the role of deubiquitinating enzymes (DUBs) in this context remains largely elusive. DUBs are essential for cells to rescue proteins from ubiquitin-mediated degradation, and to tightly control the dynamics of ubiquitin-mediated signaling events. Many DUBs associate with E3 ubiquitin ligase complexes to fine-tune the ubiquitination status of a common substrate.
Our project aims to identify and characterize DUBs that are indispensable for the correct course of autophagy. To this end, we are employing mass spectrometry-based approaches and CRISPR/Cas gRNA reagents targeting all human DUBs. We are investigating the impact of candidate DUBs in autophagy induction, autophagosome biogenesis, and selective substrate clearance. These studies will allow a better understanding of how ubiquitination modulates selective autophagy pathways by the combined action of E3 ubiquitin ligases and DUBs.
Anja Bremm

Email: bremm@em.uni-frankfurt.de
Fax: +49 (0)69 6301 5287
Phone (office): +49 (0) 69 6301-5450
Phone (lab): +49 (0)69 6301 5414
Anja Bremm

Email: bremm@em.uni-frankfurt.de
Fax: +49 (0)69 6301 5287
Phone (office): +49 (0) 69 6301-5450
Phone (lab): +49 (0)69 6301 5414
Institute of Biochemistry II
Gustav Emden-Zentrum der Biochemie
Goethe University Frankfurt - Medical Faculty
Theodor-Stern-Kai 7 / Building 75
60590 Frankfurt am Main
Germany
Tel (office) +49 (0)69 6301 5450
Tel (lab) +49 (0)69 6301 5414
Fax: +49 (0)69 6301 5287
bremm@em.uni-frankfurt.de
All IBCII Members Contact Data





