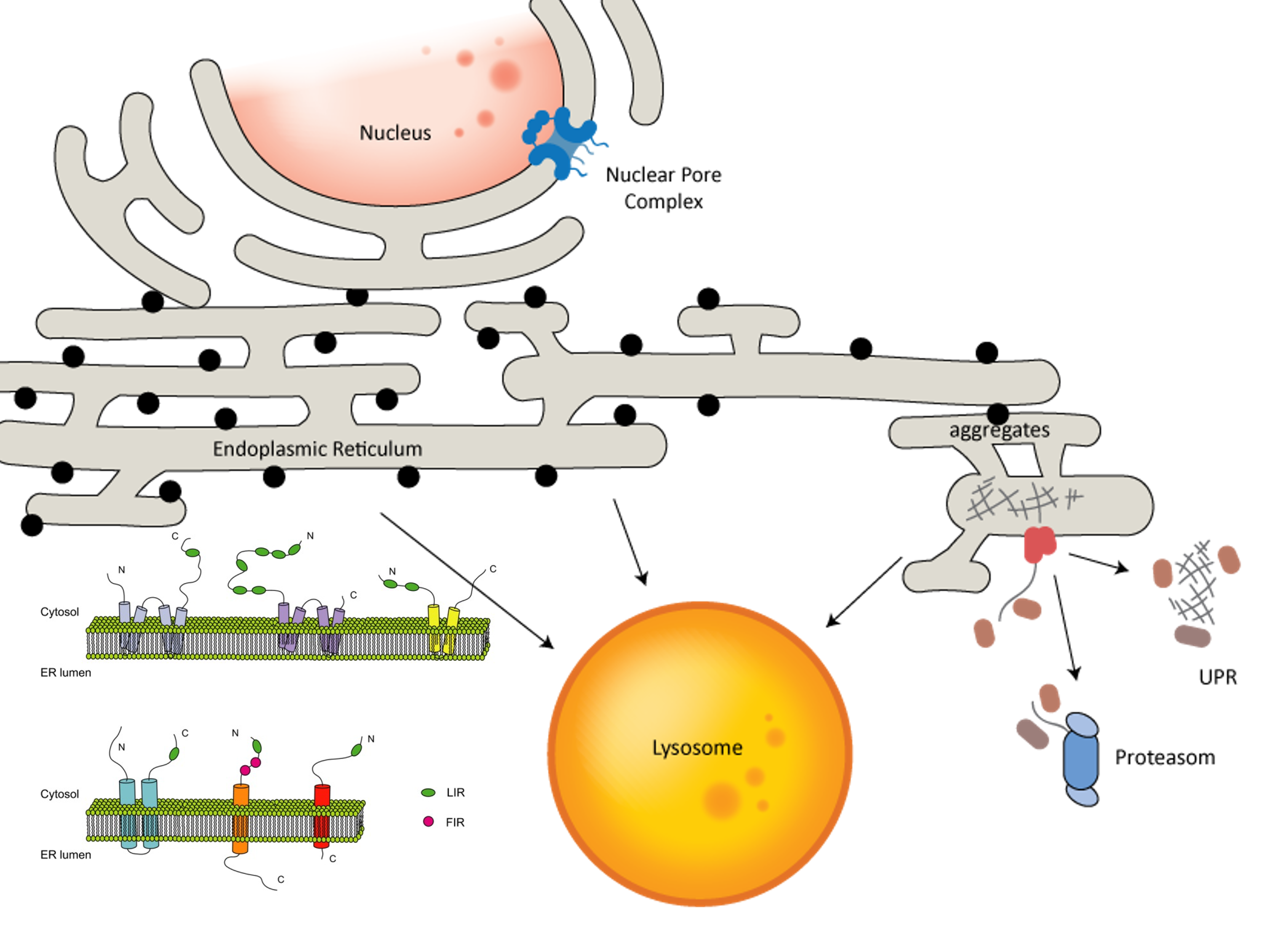
Autophagic Quality Control
Endoplasmic reticulum (ER) is a large and dynamic cellular organelle. ER morphology consists of sheets, tubules, matrixes, and contact sites shared with other membranous organelles. The capacity of the ER to fulfill its numerous biological functions depends on its continuous remodeling and the quality control of its proteome. Selective turnover of the ER membrane and ER proteins by autophagy and the proteasome, termed ER-phagy and ER associated protein degradation (ERAD) respectively, play important roles in maintaining ER morphology and functions. Research in the Stolz group focuses on different aspects of ER quality control and the crosstalk between different stress resolving pathways.
We are grateful to the funders of our work: CRC 1177 on Selective Autophagy (SFB); IMI EUbOPEN / IMI 875510 / Horizon 2020; DFG WO 210/20-2; PROXIDRUGS; ENABLE; Goethe University Frankfurt
Dr. Alexandra Stolz
Alexandra Stolz studied Biochemistry in Regensburg, Germany followed by a PhD in yeast genetics and molecular biology in Stuttgart, Germany. After a postdoc (2012-2013) working on ER associated protein degradation (ERAD) – a proteasome dependent pathway, Alexandra joined the groups of Andreas Ernst and Ivan Dikic at IBC2 in Frankfurt, Germany (2013-2016) to work on autophagy. Besides contributing to the characterization of the first autophagy receptor for ER-phagy FAM134B and elaborating the role of the kinase TBK1 in mitophagy, she utilized phage display and protein engineering to develop fluorescent sensors for the central autophagy components LC3/GABARAPs. In January 2017, she joined Genentech in South San Francisco, USA as a visiting scientist where she studied the impact of oncogene-induced secretion during cancer pathogenesis. Since February 2018, Alexandra is heading the phenotypic screening platform of the Frankfurt Competence Center of Emerging Therapeutics (FCET) and since January 2021 as well the ER quality control group located at the Buchman Institute for Molecular Life Sciences (BMLS) as an indipendent group leader. Alexandra is associated with EUbOPEN, where her group aims to identify chemical inducers and inhibitors of selective autophagy pathways.
Dr Katharina Lorentzen
Katharina studied Biological Sciences at the Universität Konstanz (Germany) and obtained a Bachelor’s Degree in 2015 and a Master’s Degree in 2018. Having received a stipend from the MRC (Medical Research Council (UK)), she then moved on to the MRC Protein Phosphorylation and Ubiquitylation Unit at the University of Dundee (UK) for her PhD with Prof Dr Ian Ganley. Her PhD focused on elucidating the mechanisms of mitophagy (the degradation of mitochondria via autophagy) and targeted organelle degradation. After a short-term post-doc position in the Ganley lab following her PhD, Katharina joined the Stolz lab in 2024 as a post-doctoral researcher to work on ER-phagy and the role and regulation of different receptors in this process.
Dr. Lorène Brunello
Lorène obtained her Master degree in Neuroscience in 2017 from the university of Montpellier (France). After receiving a Labex EpigenMed PhD scholarships, she joined Dimitris Xirodimas’ lab in Montpellier. Her PhD focused on the characterization of the nuclear protein quality control in mammals and understanding how the nucleus can eliminate nuclear protein aggregates. Lorène joined the Stolz lab in April 2022 as a post-doctoral researcher in order to broaden her expertise in quality control and investigate processes of selective autophagy in mammals.
Pablo Sanz Martinez
Pablo obtained his bachelor’s degree in biotechnology on 2018 at the university of Francisco de Vitoria in Madrid (Spain). He joined Prof. Ivan Dikic’s lab to carry out his bachelor thesis on general autophagy probes and receptors. After obtaining his degree, he enrolled in the Master in Neuroscience of Universidad Autonoma de Madrid (Spain). He started his master thesis in 2019 at the laboratory of Dr. Manuel Valiente at Centro Nacional de Investigaciones Oncologicas (CNIO). He studied the latency experienced by brain metastasis initiating cells. After obtaining his master degree, he wished to dive deeper into molecular signaling and trafficking which led him to enroll, in 2021, in a PhD at the Stolz lab where he will focus on the study of ER quality control and its implications in cell biology.
Pinar Çiftci
Pınar obtained her bachelor’s degree at Istanbul Technical University, completing a double major in Molecular Biology and Genetics as well as Chemical Engineering. She pursued her master’s degree in Cellular and Molecular Medicine at Koç University, where she studied on autophagy and cancer, focusing on developing autophagy-targeting chimeras. After obtaining her master’s degree, she joined the Stolz lab in January 2025 as a PhD student, where she is focusing on the regulation of ER-phagy and the protein complexes mediating ER interactions with other organelles.
Sara Cano
Sara was studying Biotechnology at the University Francisco de Vitoria, Spain and joined the Stolz lab in September 2020 for her Bachelor thesis on the impact of kinase signaling comparing 2D vs. 3D cultured cells. After finishing her Bachelor, Sara stayed in the lab as a research assistant and Lab Manager with a focus on cellular screens.
Sayan Biswas
Sayan completed his bachelor's in Biochemistry at the Asutosh College in Kolkata,West Bengal, India. In 2023 he completed his Master's Degree in Biochemistry from the University of Calcutta, Kolkata, West Bengal, India. During his Master’s, he worked on a dissertation project trying to characterize the physico-chemical properties of potent anti-cancer homeopathic drugs. In 2024, he joined Dr. Somdeb Bose Dasgupta’s lab as a project trainee, investigating pathogenesis and host-pathogen interactions. In 2026, he joined the Stolz lab on an SFB1177 scholarship to study regulatory mechanisms and dynamics of ER-phagy receptor clustering.
Sevi Faria
Sevi completed her Bachelor's and Master's degrees in Biochemistry at Goethe University Frankfurt, Germany. Her Bachelor's thesis, under the supervision of Prof. Dr. Zoltán Ivics, was conducted at the Paul-Ehrlich-Institute, where she applied CRISPR-Cas technology to introduce mutations in the snRNP70 gene in human cells. She completed her Master's thesis under Prof. Dr. Robert Tampé at Goethe University, where she investigated T-cell receptor clustering and T-cell activation on microstructured matrices.
Sevi joined the Stolz lab in September 2024 as a research assistant, where she supports the PROXIDRUGS.2 team in conducting research, and assisting with various molecular biology and biochemistry techniques.
Past Members:
Adrian Gackstatter
Adrian obtained his master’s degree in Molecular Biology and Neurobiology at TU Kaiserslautern In 2019, working on the impact of impaired mitochondrial protein import in D. melanogaster. Joining the Stolz Lab in 2020 as a PhD student, Adrian shifted his research interest towards the ER as part of a collaboration with the Lemberg lab at Universität zu Köln. There, he is investigating the impact of rhomboid pseudoproteases on membrane protein degradation in S. cerevisiae. Within the project, Adrian subsequently joined the Lemberg lab, to complete his project and PhD at the Universtät zu Köln.
Hassan Elsafty
Hassan studied physiology and genetics at National University Ireland Galway (NUIG), graduating with a BSC in Biomedical Science. He later completed his Masters in Biomedical Science with a focus on microbiology and infectious disease at Kingston Unversity London in 2016. His thesis investigated the isolation and determination of anti-microbial sensitivity of presumptive Campylobacter spp. with investigation of biofilm formation and microbial susceptibility to natural products, olive leaf extract and catechin hydrate.
Hassan then worked as a medical lab technician with the specialist children’s care hospital , Great Ormond Street Hospital (2015-2020) in different roles before moving to Frankfurt in 2020. He was part of the Stolz lab as a research assistant where he focused on aiding the PROXIDRUG team (www.proxidrugs.de).
Dr. Hung Ho-Xuan
Hung obtained his Bachelor's degree in Biochemistry in 2009 from the Talented Program of Hanoi University of Science, Vietnam. After that, he joined Vu Tan Trao lab as a research assistant at the National Institute of Hygiene and Epidemiology in Hanoi, Vietnam. With support from the Swiss Federal Fellowship, he completed his Master's degree in Biochemistry from the Department of Biology in Ari Helenius' lab at ETH Zürich, Switzerland. In 2013, he visited the Michael Veit lab at Free University in Berlin, Germany as a research assistant to work on the mechanism of virus egression from the cells. Starting 2014, he received the Marie Skłodowska-Curie Fellowship ‘’RNAtrain’’ to pursue his PhD in RNA Biochemistry at Gunter Meister lab at the University of Regensburg, Germany. After finishing his PhD thesis in 2018, he stayed in the lab as a postdoctoral fellow. During his time in Regensburg, he focused on analyzing the coding potential of non-coding RNAs, including circRNAs and lncRNAs, and the utilizing these microproteins’ functions in different cellular contexts. In June 2021, he joined the Stolz lab to study different aspects of selective autophagy during cellular homeostasis. In spring 2025 Hung moved to the group of Ivan Dikic for a second post doc.
Katarina Zivkovic
After studying Biology at the Belgrad University, Serbia, Katarina finished her bachelor degree in molecular biology and subsequent master degree in biomedicine at the Johannes Gutenberg University Mainz, Germany. In 2018 Katarina joined the group for 7 months on a qualification scholarship working on the establishment cell based autophagy assays before moving on to an industrial position.
Lautaro Natali
Lautaro Natali holds a Biology degree from Universidad Nacional de Córdoba (Argentina), where he graduated with honors in 2020. He conducted his bachelor's thesis at the Laboratory of Epigenetics and Cell Differentiation in the Mercedes and Martin Ferreyra Institute for Medical Research in Córdoba (Argentina), focusing on the role of the transcription factor SLUG in miRNA modulation during smooth muscle cell phenotypic switch. He subsequently obtained a PhD scholarship from the Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET) in 2020 to explore the role of RNA methylation in smooth muscle cell differentiation under the supervision of Dr. Melina Musri. Supported by the SFB1177, DAAD, and an Institute scholarship Lautaro stayed for in total 1.5 years at the Stolz lab, where he was studying the role of autophagy in this process.
Miles Willoughby
Miles completed his bachelor's degree in biochemistry at the Goethe-University of Frankfurt, Germany, under the supervision of Prof. Pos, working on heavy metal transport systems in bacteria. Due to an increasing interest in cellular biology and autophagy regulation, he joined the Stolz lab where he is investigating the regulation of ER membrane contact sites under ER stress for his master thesis. After leaving the Stolz lab, Miles started a PhD in the lab of Ivan Dikic.
Pablo Sanz Martinez
Pablo joined the Dikic group in 2018 during his last year of his biotechnology degree at Universidad Francisco de Vitoria (Madrid), to develop his bachelor thesis and lab experience. He was co-mentored by Alexandra snd stayed for 7 months working on the establishment of cell-based autophagy assays as well as the characterization of different interactors of autophagy receptors. Pablo later enrolled in the Master of Neuroscience of the Autonoma University of Madrid and made his Master thesis at the Spanish National Cancer Research Center (CNIO).
Palak Sharma
Palak completed her bachelor's in life sciences at the Sophia College for Women in Mumbai, India, working on the production of biodegradable bioplastic. In 2021 Palak moved to Germany for pursuing a Master in Physical Biology of Cells and Cell Interactions at the Goethe University Frankfurt. After an internship in the Stolz lab, Palak stayed for completing her master thesis, working in collaboration with Hung Ho-Xuan to further characterize players of a selective autophagy pathway.
Paloma Cabrerizo Poveda
Paloma obtained both a Biotechnology degree and a diploma of Expert in Methodology & Experimental Innovation during her time at the University of Francisco de Vitoria, Spain. At this time she was also a laboratory assistant under the supervision of Prof. Lourdes Rufa working in the field of Pharmacology and Pharmacognosy. For her Bachelor thesis, Paloma joined the laboratory of Prof. Vicente Rubio and Dr. Pablo Pulido at the National Center for Biotechnology (CNB-CSIC), Spain, where she worked in the area of plant molecular genetics. In September 2022 Paloma joined the Stolz group as a reseache assistant. After two years supporting the PROXIDRUGs projects, Paloma moved on to an industry postion.
Rayene Berkane
Rayene is originally from Constantine, Algeria. She obtained her first Mater’s degree in human genetics with honor from the University Constantine. In Sep. 2017, she moved to France where she obtained a second Masters in Cancer Biology from Montpellier University. In Feb. 2020, Rayene moved to Frankfurt and joined the Stolz group as a Master’s intern. In her project, she focused mainly on the establishment of drug screening assays in lung cancer cell lines cultured in 2D and 3D, respectively. After completing her Master’s thesis, Rayene intended to explore another horizon and proceeded as a PhD student, shifting her focus towards deciphering the molecular mechanism of ER quality control check mechanism around the FAM134 family. Upon successful completion of her PhD, Rayene became a postdoc at BioMedX, discovering new strategies to enhance the immonogenicity of tumors.

Sandra Bello
Sandra joined the lab in 2020 for her bachelor thesis, focusing on the impact of selected small molecules on general autophagy and ER-phagy flux, respectively.
Tristan Gläsner
Tristan studies Bioscienes – Applied Biology for Medicine and Pharmacy at the Fresenius University of Applied Sciences. In his 5th semester he joined the Stolz lab for 6 months of wet-lab training. During his stay he is working with Rayene Berkane on the post-translational cues regulating the activity of FAM134 proteins. After working in the lab as as HiWi, Tristan started his Bachelor Thesis in February 2023.
Ho-Xuan H, Bruckmann A, Natali L, Prieto-Garcia C, Stuke JFM, Brunello L, Vicente A, Pfab A, Wesseling H, Cano-Franco S, Khatri A, Larivera S, Sanz-Martinez P, de la Cruz-Thea B, Jacomin AC, Prochazka J, Feederle R, Dötsch V, Sedlacek R, Hummer G, Kaiser S, Dikic I, Musri MM, Meister G, Stolz A, et al. YTHDF proteins and mA-RNA clients undergo autophagic turnover during contact inhibition. Cell Rep 2025. 44 (9) 116188 Link
Tan EP, Lyang N, Doroodian S, Sanz-Martinez P, Xu J, Zaretski S, Nieto-Torres JL, Ebata H, Lim SHY, Hou WC, Clay KJ, Yoon L, Massey LA, Rhoades D, Garza D, Johnson KA, To A, Ambaye L, Bentley EP, Petrascheck M, Stolz A, Kelly JW, Hansen M Autophagy activator AA-20 improves proteostasis and extends lifespan. Proc Natl Acad Sci U S A 2025. 122 (32) e2423455122 Link
Buonomo V, Lohachova K, Reggio A, Cano-Franco S, Cillo M, Santorelli L, Venditti R, Polishchuk E, Peluso I, Brunello L, Cirillo C, Petrosino S, Silva M, De Cegli R, Di Bartolomeo S, Gargioli C, Swuec P, Cortese M, Stolz A, Bhaskara RM, Grumati P Two FAM134B isoforms differentially regulate ER dynamics during myogenesis. EMBO J 2025. 44 (4) 1039-1073 Link
Tredup C, Ackloo S, Beck H, Brown PJ, Bullock AN, Ciulli A, Dikic I, Edfeldt K, Edwards AM, Elkins JM, Farin HF, Fon EA, Gstaiger M, Günther J, Gustavsson AL, Häberle S, Isigkeit L, Huber KVM, Kotschy A, Krämer O, Leach AR, Marsden BD, Matsui H, Merk D, Montel F, et al. Toward target 2035: EUbOPEN - a public-private partnership to enable & unlock biology in the open. RSC Med Chem 2025. 16 (2) 457-464 Link
Schwalm MP, Dopfer J, Kumar A, Greco FA, Bauer N, Löhr F, Heering J, Cano-Franco S, Lechner S, Hanke T, Jaser I, Morasch V, Lenz C, Fearon D, Marples PG, Tomlinson CWE, Brunello L, Saxena K, Adams NBP, von Delft F, Müller S, Stolz A, Proschak E, Kuster B, Knapp S, et al. Critical assessment of LC3/GABARAP ligands used for degrader development and ligandability of LC3/GABARAP binding pockets. Nat Commun 2024. 15 (1) 10204 Link
Prieto-Garcia C, Matkovic V, Mosler T, Li C, Liang J, Oo JA, Haidle F, Mačinković I, Cabrera-Orefice A, Berkane R, Giuliani G, Xu F, Jacomin AC, Tomaskovic I, Basoglu M, Hoffmann ME, Rathore R, Cetin R, Boutguetait D, Bozkurt S, Hernández Cañás MC, Keller M, Busam J, Shah VJ, Wittig I, et al. Pathogenic proteotoxicity of cryptic splicing is alleviated by ubiquitination and ER-phagy. Science 2024. 386 (6723) 768-776 Link
Sanz-Martinez P, Berkane R, Stolz A Function of CSNK2/CK2 selectively affects the endoplasmic reticulum and the Golgi apparatus in mtor-mediated autophagy induction. Autophagy 2025. 21 (2) 480-486 Link
Isigkeit L, Schallmayer E, Busch R, Brunello L, Menge A, Elson L, Müller S, Knapp S, Stolz A, Marschner JA, Merk D Chemogenomics for NR1 nuclear hormone receptors. Nat Commun 2024. 15 (1) 5201 Link
Sanz-Martinez P, Tascher G, Cano-Franco S, Cabrerizo-Poveda P, Münch C, Homan EJ, Stolz A GPCR Function in Autophagy Control: A Systematic Approach of Chemical Intervention. J Mol Biol 2024. 436 (15) 168643 Link
Stolz A CSNK2/CK2 regulates selective autophagy of the endoplasmic reticulum. Autophagy 2024. 20 (7) 1694-1695 Link
Berkane R, Ho-Xuan H, Glogger M, Sanz-Martinez P, Brunello L, Glaesner T, Kuncha SK, Holzhüter K, Cano-Franco S, Buonomo V, Cabrerizo-Poveda P, Balakrishnan A, Tascher G, Husnjak K, Juretschke T, Misra M, González A, Dötsch V, Grumati P, Heilemann M, Stolz A The function of ER-phagy receptors is regulated through phosphorylation-dependent ubiquitination pathways. Nat Commun 2023. 14 (1) 8364 Link
Nalbach K, Schifferer M, Bhattacharya D, Ho-Xuan H, Tseng WC, Williams LA, Stolz A, Lichtenthaler SF, Elazar Z, Behrends C Author Correction: Spatial proteomics reveals secretory pathway disturbances caused by neuropathy-associated TECPR2. Nat Commun 2023. 14 (1) 8322 Link
Sharma R, Adams M, Griffith-Jones S, Sahr T, Gomez-Valero L, Weis F, Hons M, Gharbi S, Berkane R, Stolz A, Buchrieser C, Bhogaraju S Structural basis for the toxicity of Legionella pneumophila effector SidH. Nat Commun 2023. 14 (1) 7068 Link
Cano-Franco S, Ho-Xuan H, Brunello L, Stolz A Live-Cell High-Throughput Screen for Monitoring Autophagy Flux. Methods Mol Biol 2023. 2706 215-224 Link
Nalbach K, Schifferer M, Bhattacharya D, Ho-Xuan H, Tseng WC, Williams LA, Stolz A, Lichtenthaler SF, Elazar Z, Behrends C Spatial proteomics reveals secretory pathway disturbances caused by neuropathy-associated TECPR2. Nat Commun 2023. 14 (1) 870 Link
Di Lorenzo G, Iavarone F, Maddaluno M, Plata-Gómez AB, Aureli S, Quezada Meza CP, Cinque L, Palma A, Reggio A, Cirillo C, Sacco F, Stolz A, Napolitano G, Marin O, Pinna LA, Ruzzene M, Limongelli V, Efeyan A, Grumati P, Settembre C Phosphorylation of FAM134C by CK2 controls starvation-induced ER-phagy. Sci Adv 2022. 8 (35) eabo1215 Link
Michel M, Benítez-Buelga C, Calvo PA, Hanna BMF, Mortusewicz O, Masuyer G, Davies J, Wallner O, Sanjiv K, Albers JJ, Castañeda-Zegarra S, Jemth AS, Visnes T, Sastre-Perona A, Danda AN, Homan EJ, Marimuthu K, Zhenjun Z, Chi CN, Sarno A, Wiita E, von Nicolai C, Komor AJ, Rajagopal V, Müller S, et al. Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function. Science 2022. 376 (6600) 1471-1476 Link
Hammer A, Cerretti G, Ricciardi DA, Schiffmann D, Maranda S, Kummer R, Zumbühl C, Rattenbacher-Kiser KF, von Arx S, Ammann S, Strobl F, Berkane R, Stolz A, Stelzer EHK, Egli M, Schleiff E, Wuest SL, Böhmer M Retrograde Analysis of Calcium Signaling by CaMPARI2 Shows Cytosolic Calcium in Chondrocytes Is Unaffected by Parabolic Flights. Biomedicines 2022. 10 (1) Link
Basic M, Hertel A, Bajdzienko J, Bonn F, Tellechea M, Stolz A, Kern A, Behl C, Bremm A The deubiquitinase USP11 is a versatile and conserved regulator of autophagy. J Biol Chem 2021. 297 (5) 101263 Link
Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cadwell K, Cecconi F, Choi AMK, Choi ME, Chu CT, Codogno P, Colombo MI, Cuervo AM, Deretic V, Dikic I, Elazar Z, Eskelinen EL, Fimia GM, Gewirtz DA, Green DR, Hansen M, Jäättelä M, Johansen T, et al. Autophagy in major human diseases. EMBO J 2021. 40 (19) e108863 Link
Reggio A, Buonomo V, Berkane R, Bhaskara RM, Tellechea M, Peluso I, Polishchuk E, Di Lorenzo G, Cirillo C, Esposito M, Hussain A, Huebner AK, Hübner CA, Settembre C, Hummer G, Grumati P, Stolz A Role of FAM134 paralogues in endoplasmic reticulum remodeling, ER-phagy, and Collagen quality control. EMBO Rep 2021. 22 (9) e52289 Link
Wells CI, Al-Ali H, Andrews DM, Asquith CRM, Axtman AD, Dikic I, Ebner D, Ettmayer P, Fischer C, Frederiksen M, Futrell RE, Gray NS, Hatch SB, Knapp S, Lücking U, Michaelides M, Mills CE, Müller S, Owen D, Picado A, Saikatendu KS, Schröder M, Stolz A, Tellechea M, Turunen BJ, et al. The Kinase Chemogenomic Set (KCGS): An Open Science Resource for Kinase Vulnerability Identification. Int J Mol Sci 2021. 22 (2) Link
Zielke S, Kardo S, Zein L, Mari M, Covarrubias-Pinto A, Kinzler MN, Meyer N, Stolz A, Fulda S, Reggiori F, Kögel D, van Wijk S ATF4 links ER stress with reticulophagy in glioblastoma cells. Autophagy 2021. 17 (9) 2432-2448 Link
Shin D, Mukherjee R, Liu Y, Gonzalez A, Bonn F, Liu Y, Rogov VV, Heinz M, Stolz A, Hummer G, Dötsch V, Luo ZQ, Bhogaraju S, Dikic I Regulation of Phosphoribosyl-Linked Serine Ubiquitination by Deubiquitinases DupA and DupB. Mol Cell 2020. 77 (1) 164-179.e6 Link
Stolz A, Grumati P The various shades of ER-phagy. FEBS J 2019. 286 (23) 4642-4649 Link
Chaikuad A, Koschade SE, Stolz A, Zivkovic K, Pohl C, Shaid S, Ren H, Lambert LJ, Cosford NDP, Brandts CH, Knapp S Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2). Biochem J 2019. 476 (5) 875-887 Link
Rogov VV, Stolz A, Ravichandran AC, Rios-Szwed DO, Suzuki H, Kniss A, Löhr F, Wakatsuki S, Dötsch V, Dikic I, Dobson RC, McEwan DG Structural and functional analysis of the GABARAP interaction motif (GIM). EMBO Rep 2018. 19 (12) Link
Stolz A, Dikic I Elusive mitochondrial connection to inflammation uncovered. Nature 2018. 561 (7722) 185-186 Link
Grumati P, Dikic I, Stolz A ER-phagy at a glance. J Cell Sci 2018. 131 (17) Link
Stolz A, Dikic I Heterotypic Ubiquitin Chains: Seeing is Believing. Trends Cell Biol 2018. 28 (1) 1-3 Link
Rogov VV, Stolz A, Ravichandran AC, Rios-Szwed DO, Suzuki H, Kniss A, Löhr F, Wakatsuki S, Dötsch V, Dikic I, Dobson RC, McEwan DG Structural and functional analysis of the GABARAP interaction motif (GIM). EMBO Rep 2017. 18 (8) 1382-1396 Link
Stolz A, Putyrski M, Kutle I, Huber J, Wang C, Major V, Sidhu SS, Youle RJ, Rogov VV, Dötsch V, Ernst A, Dikic I Fluorescence-based ATG8 sensors monitor localization and function of LC3/GABARAP proteins. EMBO J 2017. 36 (4) 549-564 Link
Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, Youle RJ, Dikic I Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A 2016. 113 (15) 4039-44 Link
Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, Mauthe M, Katona I, Qualmann B, Weis J, Reggiori F, Kurth I, Hübner CA, Dikic I Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 2015. 522 (7556) 354-8 Link
Stolz A, Dikic I PINK1-PARKIN interplay: down to ubiquitin phosphorylation. Mol Cell 2014. 56 (3) 341-342 Link
Stolz A, Ernst A, Dikic I Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 2014. 16 (6) 495-501 Link
Stolz A, Besser S, Hottmann H, Wolf DH Previously unknown role for the ubiquitin ligase Ubr1 in endoplasmic reticulum-associated protein degradation. Proc Natl Acad Sci U S A 2013. 110 (38) 15271-6 Link
Stolz A, Wolf DH Use of CPY and its derivatives to study protein quality control in various cell compartments. Methods Mol Biol 2012. 832 489-504 Link
Martinez Benitez E, Stolz A, Becher A, Wolf DH Mnl2, a novel component of the ER associated protein degradation pathway. Biochem Biophys Res Commun 2011. 414 (3) 528-32 Link
Wolf DH, Stolz A The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim Biophys Acta 2012. 1823 (1) 117-24 Link
Benitez EM, Stolz A, Wolf DH Yos9, a control protein for misfolded glycosylated and non-glycosylated proteins in ERAD. FEBS Lett 2011. 585 (19) 3015-9 Link
Stolz A, Hilt W, Buchberger A, Wolf DH Cdc48: a power machine in protein degradation. Trends Biochem Sci 2011. 36 (10) 515-23 Link
Stolz A, Schweizer RS, Schäfer A, Wolf DH Dfm1 forms distinct complexes with Cdc48 and the ER ubiquitin ligases and is required for ERAD. Traffic 2010. 11 (10) 1363-9 Link
Stolz A, Wolf DH Endoplasmic reticulum associated protein degradation: a chaperone assisted journey to hell. Biochim Biophys Acta 2010. 1803 (6) 694-705 Link
ER network integrity and turnover by autophagy rely on specific ER-phagy receptors, which influence and coordinate alterations in ER morphology and the degradation of ER contents and membranes via the lysosome, by interacting with the LC3/GABARAP family. The first mammalian ER-phagy receptor, FAM134B, was identified in our institute. My laboratory is specifically interested in its two family members - FAM134A and FAM134C – in potential overlapping and distinct functions as well as their crosstalk and regulation.
The ERAD process is organized around ER resident, membrane-embedded E3 ubiquitin ligases forming distinct protein complexes and collectively providing the essential substrate ubiquitination activity for degradation of ERAD substrates by the proteasome. Recent high-resolution structures revealed that the yeast E3 ligase Hrd1 and the rhomboid pseudoprotease Der1 provide a sizable pore for the dislocation of luminal proteins into the cytoplasm. In collaboration with Marius Lemberg and Dönem Avci (Uni Heidelberg), we aim to define the precise physiological role of Dfm1 in S. cerevisae, identify endogenous substrates, and decipher the molecular mechanism of membrane protein degradation in the ER – potentially linking ERAD and ER-phagy
doi: 10.1111/febs.15031
DOI: 10.15252/embr.201643587
DOI: 10.1038/nature14498
DOI: 10.1111/j.1600-0854.2010.01093.x
We utilize microscopy instruments (Sartorius IncuCyte S3 and Yokogawa CQ1) to perform cell-based assays. Depending on the scientific question, we use a variety of fluorescent labeled reporter cell lines and compound libraries to identify signaling pathways or study the impact of specific stress and pathway alterations on cell proliferation. To allow semi-high throughput our assays are mostly performed in 384-well plate format, including triplicates of each tested condition as well as standard control compounds on each plate for comparison and quality control. We are able to precisely handle minimal amount of compounds with the help of Labcyte’s ECHO 555 liquid handler.
Part of our efforts is channeled into the establishment of spheroid based 3D assays. Their remarkable clinical relevance and realistic setting in respect of cellular behavior and pharmacological response compared to 2D cell culture make them powerful tools for biologic studies and drug development.
DOI: 10.1042/BCJ20190038
bioRxiv (dx.doi.org/10.1101/2019.12.22.886523)
BMLS
Alexandra Stolz

Email: stolz@em.uni-frankfurt.de
Fax: +49 (0) 69 6301 5577
Phone: +49 (0) 69 798-42589
Alexandra Stolz

Email: stolz@em.uni-frankfurt.de
Fax: +49 (0) 69 6301 5577
Phone: +49 (0) 69 798-42589
All IBCII Members Contact Data






















