23 Aug 2025 - Autophagy tunes m⁶A reader proteins during contact inhibition
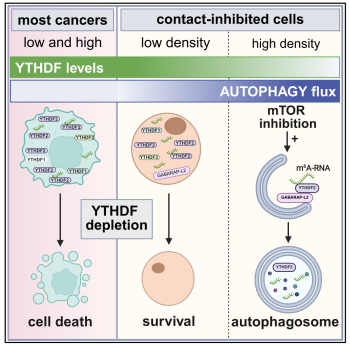
Scientists from IBC2 in collaboration with colleagues from the University of Regensburg show that when non-transformed cells reach high density and contact inhibition kicks in, the m⁶A reader proteins YTHDF1-3 are actively cleared by autophagy, a process that can also be triggered with the mTOR inhibitor Torin1. In contrast, cancer cells that evade contact inhibition retain YTHDF proteins, pointing to a growth-control checkpoint that tumors bypass. The work, led by Alexandra Stolz and Gunter Meister as well as Hung Ho-Xuan as the leading author, links YTHDF stability directly to the mTOR–autophagy axis and suggests coordinated turnover of both proteins and their m⁶A-modified RNA clients by selective autophagy.
“Contact inhibition doesn’t just pause proliferation - it rewires cellular quality control,” says Stolz. “Our data indicate that YTHDF proteins carry conserved LIR motifs that can be activated and connect to autophagic membrane; together with previously reported proteasomal degradation, this creates a tightly tuned system that adjusts YTHDF abundance and RNA fate according to growth cues.” Ho-Xuan adds, “In preclinical models, lowering YTHDF levels, especially YTHDF2, compromised cancer cell survival, while non-transformed cells seem to be largely unaffected. It’s an intriguing vulnerability that merits careful follow-up.”
The authors emphasize that the findings are based on laboratory and xenograft studies and that key questions remain, including which RNAs are preferentially targeted and how broadly this mechanism operates across biological contexts. Future work will map upstream signals (including phosphorylation near LIR motifs), define RNA cargoes routed to autophagosomes, and assess whether modulating this interface can influence cell-state transitions or expose therapeutic opportunities.