10 May 2012 - A novel mechanism regulating SUMO-dependent protein networks.
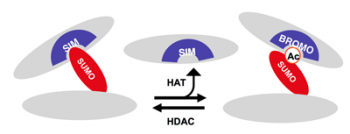
The covalent attachment of the ubiquitin-like modifier SUMO to proteins serves an important mechanism for the control of protein-protein interactions. This is generally mediated via recognition of a SUMO-conjugate by an interaction partner that contains a specific SUMO interaction module, termed SIM (SUMO interaction motif). A major question is how the dynamics of SUMO/SIM interactions are regulated.
Recent work done by Rebecca Ullmann in Stefan Muller's group together with co-workers from the Lombardi Cancer Center in Washington now uncovered an acetyl-dependent switch that determines the selectivity and dynamics of SUMO-SIM interactions (Molecular Cell, online May 10, 2012).
In this context they show that acetylation of SUMO within a basic interface prevents binding to SIMs in PML, Daxx, and PIAS. One the other hand, acetyl-SUMO specifically binds to the Bromodomain of the co-activator p300. This acetyl-dependent switch of SUMO-mediated protein interactions attenuates SUMO-regulated gene silencing and affects the assembly of PML nuclear bodies. This work thus unravels a novel interplay of post-translational modifications that expands the regulatory repertoire of SUMO signaling.
Original Article:
Ullmann R, Chien C D, Avantaggiati M L, Muller S (2012) An acetylation switch regulates SUMO-dependent protein interaction networks, Mol Cell, online May 10, 2012, DOI 10.1016/j.molcel.2012.04.006