News from the Institute

Christian Münch, group leader at the Institute of Biochemistry II of Goethe University Frankfurt, was selected as an EMBO Young Investigator. The EMBO Young Investigator Program recognizes and supports life scientists, who have already demonstrated scientific excellence and are within the first years of their independent group. It provides with financial support, access to the European Molecular Biology Laboratory (EMBL) core facilities, as well as a variety of training and networking opport
... (read more)
The Academy of Sciences and Literature in Mainz has welcomed 13 new members in their ranks, among them IBC2 Director Ivan Đikić. Đikić was in prominent company – BioNTech founders Uğur Şahin and Özlem Tureci were also amongst the new members announced. The Mainz Academy is one of Germany’s eight academies of science, its objectives are the cultivation of sciences and literature as well as preservation and promotion of culture. It is divided into three Classes: Mathematics and Natural Sciences, Humanities and Social Sciences, Literature and Music.
... (read more)
The list of Highly Cited Researcher identifies pioneers in their respective fields who repeatedly demonstrated significant and broad influence. IBC2 Director Ivan Đikić is honored for generating exceptional impact by publishing multiple highly cited papers in the past decade in even two fields, “Biology and Biochemistry” and “Molecular Biology and Genetics”. His papers in these fields rank in the top 1% by citations. The analysis is based on citations as recorded in Web of Science, for 2021 all publications between 2010 and 2020 were considered.
... (read more)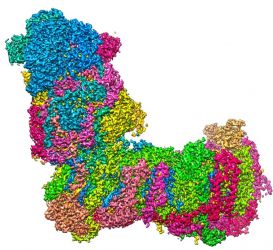
All life processes require a constant supply of energy. To meet this demand, ATP serves as the cell's molecular fuel. The driving force for ATP synthesis is the so-called mitochondrial respiratory chain, with mitochondrial complex I forming the starting point. Mitochondrial complex I is a very large membrane protein complex, but despite its central role in aerobic energy metabolism, its molecular mechanism is still largely unknown. In a study published today in Science Advances, the Zickermann group together with colleagues from the MPI of Biophysics and the University of Helsinki now provides groundbreaking new insights into this key player of biological energy conversion.
... (read more)
IBC2 EMBO-Postdoctoral Fellow Dr. Santosh Kumar Kuncha was awarded the INSA Medal For Young Scientists by the Indian National Science Academy. The INSA Medal is one of the highest recognition for young researchers in India.
... (read more)