30 Dec 2025 - Comprehensive map of human E3 ligases reveals new opportunities for targeted protein degradation
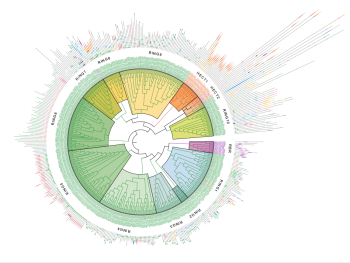
E3-Ligome: Researchers at Goethe University have elucidated the relationships among all 462 catalytic human E3 ligases.
Image: Ramachandra M. Bhaskara, Goethe University Frankfurt
In a new publication in Nature Communications, a team of researchers from IBC2 at Goethe University Frankfurt, reports the first comprehensive, data-driven classification of all human E3 ubiquitin ligases. The study presents a systematic map of the human “E3 ligome” and provides new insights into the functional relationships among E3 ligases.
Using AI-supported computational analyses combined with experimental validation, the researchers classified E3 ligases into distinct families and identified 40 E3 ligases with potential relevance for targeted protein degradation strategies such as PROTACs. As E3 ligases act as key mediators in the ubiquitin–proteasome system, this expanded view of the E3 ligase landscape opens new opportunities to diversify proximity-inducing drug design beyond the few E3 ligases currently in use.
The complete E3 ligome dataset has been made publicly available, providing an important resource for the research community and supporting the development of next-generation proximity-inducing therapeutics.