01 Dec 2016 - New ubiquitin chemistry regulates life processes.
In the latest issue of Cell, a team around IBC2 director Ivan Dikic reveals molecular details of a novel ubiquitination mechanism that may affect numerous life processes.
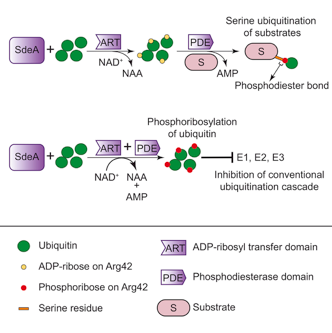
Earlier this year, U.S. colleagues reported that Legionella enzyme SdeA is capable of catalysing ubiquitination single-handedly. Now, the Frankfurt scientists together with collaborators from the MPI for Biology of Ageing (Cologne) have elucidated the chemistry behind and discovered a hitherto unknown type of linkage between ubiquitin and target proteins.
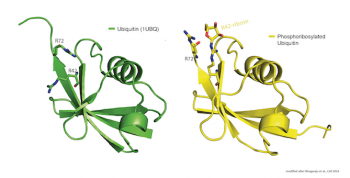
Unlike the conventional ubiquitination reaction, the novel one is NAD-dependent, involving an ADP-ribose intermediate and resulting in the attachment of ubiquitin to substrate serine residues via a phosphodiester bond.
While those findings alone are breaking new ground, the discovery went even further: The team showed that the Legionella enzyme does not only transfer ubiquitin onto target proteins, but also leaves behind a complete pool of chemically modified, phosphoribosylated ubiquitin.
Phosphoribosylated ubiquitin almost completely inhibits the conventional ubiquitination system and thereby affects essential cellular processes, e.g. proteasomal protein degradation, mitophagy and pro-inflammatory signalling.
This explains the pathogenic effects of Legionella infection in immunocompromised patients, who often suffer from extensive lung tissue damage despite antibiotic treatment. The insight generated by the Dikic team may now open the road to the development of new antibacterial agents, which could complement conventional antibiotics by limiting the cellular damage induced by bacterial enzymes.